Abstract
Aggregative adherence to human epithelial cells, most to renal proximal tubular (HK-2) cells, and biofilm formation was identified among antimicrobial resistant Escherichia coli strains mainly isolated from bacteremia. The importance of these virulence properties contributing to host colonization and infection associated with multiresistant E. coli should not be neglected.
adherence; biofilm; extraintestinal Escherichia coli; antimicrobial resistance; virulence genes
Escherichia coli is responsible for a wide variety of community and hospital acquired extraintestinal infections, showing increasing antimicrobial resistance rates. Extraintestinal pathogenic E. coli can inhabit the intestinal tract as part of the microflora and infect extraintestinal sites such as the urinary tract, the bloodstream, and the central nervous system (Kaper et al., 2004Kaper JB, Nataro JP, Harry LT et al. (2004) Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140.). Ability to adhere to different surfaces, and formed biofilms have been highlighted as important features associated to E. coli virulence (Boll et al., 2013Boll EJ, Struve C, Boisen N et al. (2013) Role of Enteroaggregative Escherichia coli Virulence Factors in Uropathogenesis Infect Immun 81:1164–1171.; Peirano et al., 2013Peirano G, Mulvey GL, Armstrong GD et al. (2013) Virulence potential and adherence properties of Escherichia coli that produce CTX-M and NDM β-lactamases. J Med Microbiol 62:525–530.). Additionally, resistance to antimicrobials in biofilm-forming isolates contributes to bacterial persistence which may lead to chronic infections and treatment problems. The present study aimed to determine the ability of eleven extraintestinal and one intestinal E. coli isolates, showing resistance to antimicrobials, to form biofilm and interact with different epithelial cells. These strains were previously isolated from community and hospital-acquired human infections of patients admitted to a private General Hospital in the city of São Paulo, Brazil, during 4 months in 2006, and two of them carryed blaCTX-M-14 and blaCTX-M-15 genes, codifying for CTX-M-14 and CTX-M-15 β-lactamases (Cergole-Novella et al., 2010Cergole-Novella MC, Guth BEC, Castanheira M et al.(2010) First description of bla (CTX-M-14)- and bla (CTX-M-15)-producing Escherichia coliisolates in Brazil. Microb Drug Resist 16:177–184.). Moreover, the presence of several virulence markers related to adhesins, toxins, protectins, elements involved in iron acquisition, and invasin reported to be associated with pathogenesis has been sought for. All these aspects provide additional information that may be useful for epidemiological research on these pathogenic strains.
The assay for visualization of biofilm formation was performed in glass tube according to the protocol described by Uhlich et al.(2006)Uhlich GA, Cooke PH, Solomon EB (2006) Analyses of the red-dry rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol 72:2564–2572. with some modifications. A visible pellicle which stained with crystal violet formed immediately below the surface of the medium indicating the ability to form biofilm could be observed in 8 E. coli strains after growth at 37 °C for 24 and 48 h (Table 1). Three isolates adhered to glass only when cultured in LB with salt, whereas two others formed biofilm when grown in LB without salt (data not shown). Some previous studies have shown that biofilm formation by extraintestinal E. coli can be associated with the expression of different adhesins such as curli fimbriae (Uhlich et al., 2006Uhlich GA, Cooke PH, Solomon EB (2006) Analyses of the red-dry rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol 72:2564–2572.). In order to identify the presence of gene sequences related to curli (csgA and crl) in the isolates studied, PCR assays were carried out, and curli expression and production of cellulose were determined according to Biscola et al. (2011)Biscola FT, Abe CM, Guth BEC (2011) Determination of adhesin gene sequences in, and biofilm formation by, O157 and non-O157 Shiga toxin-producing Escherichia coli strains isolated from different sources. Appl Environ Microbiol 77: 2201–2208.. Production of cellulose, a major exopolysaccharide component of the biofilm matrix, has been shown to enhance bacterial adherence (Zogaj et al., 2001Zogaj X, Nimtz M, Rohde M et al. (2001) The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463.). Although, csgA and crl genes were identified in all E. coli isolates studied, expression of curli, as judged by characteristic red-colored colonies formed on Congo red agar plates, was observed in only three biofilm-producing isolates (Table 1). E. coli isolate 43 formed both curli-positive and curli-negative colonies on Congo red agar (curli-variant strain) (Table 1). None of the E. coli curli-positive isolates produced cellulose as confirmed by the calcofluor binding assay (Biscola et al. (2011)Biscola FT, Abe CM, Guth BEC (2011) Determination of adhesin gene sequences in, and biofilm formation by, O157 and non-O157 Shiga toxin-producing Escherichia coli strains isolated from different sources. Appl Environ Microbiol 77: 2201–2208., but production of cellulose was identified in only one curli and biofilm negative isolate (Table 1). It is widely recognized that bacteria colonizing a surface as a biofilm can be much more resistant to antimicrobial chemotherapy (Stewart, 1994Stewart PS (1994) Biofilm Accumulation Model That Predicts Antibiotic Resistance of Pseudomonas aeruginosa Biofilms. Antimicrob Agents Chemother 38:1052–1058.). Multiresistance (defined as resistance to three or more antimicrobial groups) was observed in 8 of 12 (66.7%) isolates analyzed (Table 1). Six of the eight multiresistant E. coli isolates formed biofilm, including the CTX-M-producing isolates 34 and 35. Therefore, the combination of biofilm formation and resistance to multiple antimicrobials is likely to have contributed to the bacterial fitness and their potential to cause infection.
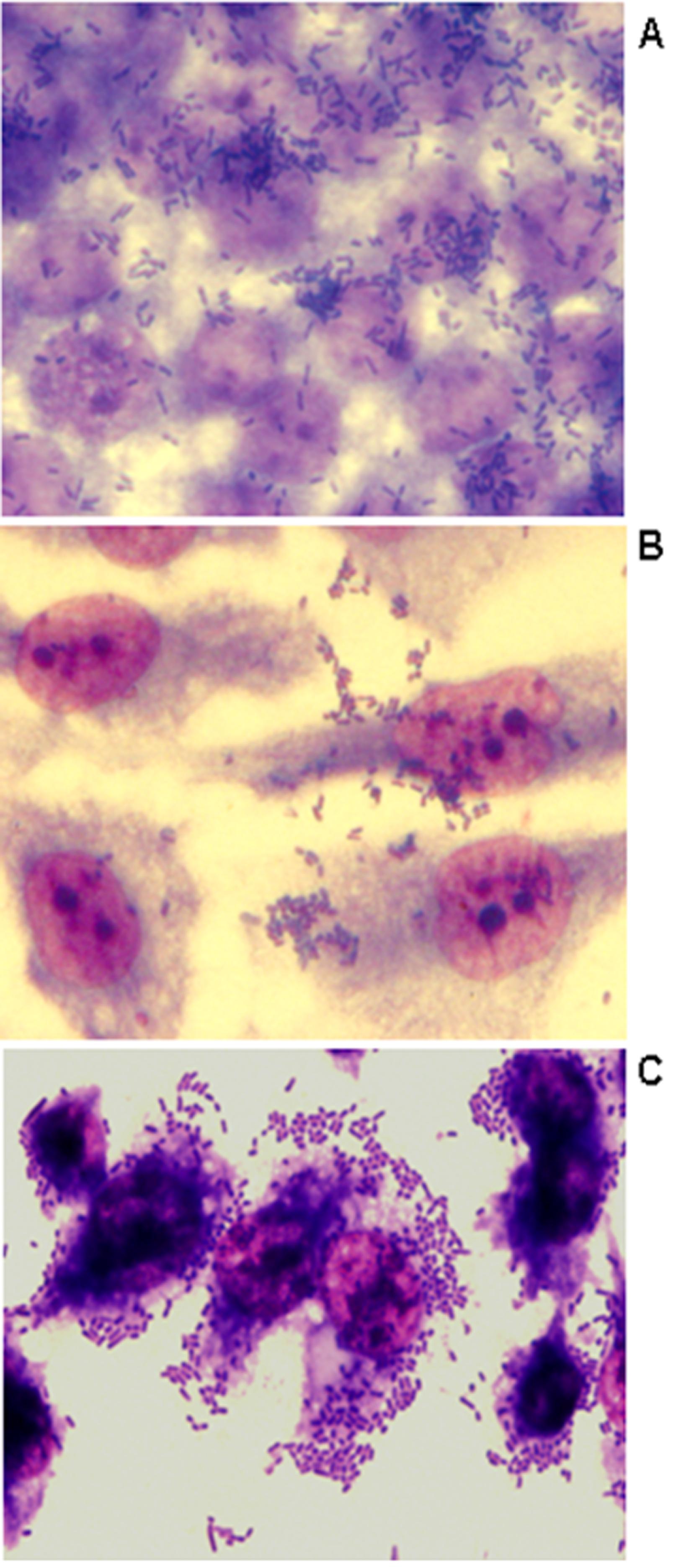
The ability of bacteria to adhere to HeLa, CaCo-2, and to human renal proximal tubular epithelial (HK-2) cells was assessed as described previously by Cravioto et al. (1979)Cravioto A, Gross RJ, Scotland SM et al. (1979) An adhesive factor found in strains of E. coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol 3:95–99.. Adherence to cells was identified in 50% to 58% of the E. coli isolates studied, confirming the importance of bacterial adherence for host colonization (Table 1 and Figure 1). Most of the isolates did not present a defined adherence pattern to CaCo-2 and to HeLa cells, except for E. coli isolate 47 that showed a diffuse adherence to HeLa cells after 3 h of incubation (data not shown). All multiresistant isolates studied presented biofilm formation and/or adherence to at least one of the cells studied. On the other hand, six of the seven isolates that adhered to HK-2 cells presented an aggregative adherence (AA) (Figure 1C), including the CTX-M-producing isolates. To our knowledge, aggregative adherence of extraintestinal E. coli isolates to HK-2 cells has not been previously described. However, some previous reports described an enteroaggregative E. coli(EAEC) isolate that was able to form an intense biofilm on urethral catheters (Boll et al., 2013Boll EJ, Struve C, Boisen N et al. (2013) Role of Enteroaggregative Escherichia coli Virulence Factors in Uropathogenesis Infect Immun 81:1164–1171.). Although showing aggregative adhesion (AA), the presence of aggR(transcriptional activator of aggregative adherence fimbriae), aatA(outer membrane protein), aggA (major subunit of the aggregative adherence fimbriae type I), and aafA (major adhesive pilin subunit of the aggregative adherence fimbriae type II) genes, molecular markers of EAEC could not be detected by PCR assays as described by Monteiro et al. (2009)Monteiro BT, Campos LC, Sircili MP et al. (2009) The dispersin-encoding gene (aap) is not restricted to enteroaggregative Escherichia coli. Diag Microbiol Infect Dis 65:81–84.. Nevertheless, the AA phenotype identified in the multiresistant extraintestinal E. coli isolates studied here can reflect an enhance ability of these isolates to adhere to eukaryotic cells.
Representative adherence to CaCo-2 (A), HeLa (B) and HK-2 (C) human epithelial cells infected with CTX-M-14-producing E. coli isolate 34 (6 h assay). Cells were subjected to Giemsa and May-Grünwald staining. Original magnification X1,000.
The presence of several virulence factors helps the organisms to adhere, colonize and/or invade host cells, and incite a noxious inflammatory response, thereby giving rise to clinical disease (Johnson et al., 2001Johnson JR, O’Bryan TT, Kuskowski M et al. (2001) Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect Immun 69:5363–5374.). In order to evaluate the ability of these bacteria to invade CaCo-2 cells, invasion assays were performed as described previously by Robins-Browne and Bennett-Wood (1992)Robins-Browne RM, Bennett-Wood V (1992) Quantitative assessment of the ability of Escherichia coli to invade cultured animal cells. Microb Pathog 12:159–164. with some modifications. This assay was the same as that for adhesion, except that after incubation for 3 h and removal of non-adherent bacteria by washing, cells were further incubated in Dulbecco’s Modified Eagle Medium/Ham’s F-12 (DMEM/F-12) containing gentamicin or kanamicin. A low invasion index of 0 up to 2% was observed for most of the E. coli isolates studied (Table 1) when compared to the positive control E. coli EH41 (7.6%). However, higher invasion indexes were observed for E. coliisolates 35, 36, 40 and 44. As a matter of interest CTX-M-producing E. coli isolates 34 and 35, obtained respectively from the feces and blood culture from the same patient, and presenting identical serotype, antimicrobial susceptibility profile and sharing more than 90% of similarity by PFGE analysis (Cergole-Novella et al., 2010Cergole-Novella MC, Guth BEC, Castanheira M et al.(2010) First description of bla (CTX-M-14)- and bla (CTX-M-15)-producing Escherichia coliisolates in Brazil. Microb Drug Resist 16:177–184.), presented different invasion abilities with the highest index being observed for the isolate that was recovered from the blood culture. One can suggest that this higher invasion ability may have contributed to the persistence of this particular isolate in the host.
In an attempt to analyze the virulence panel carried by the extraintestinal E. coli isolates studied, presence of P fimbriae (pyelonephritis-associated pilus encoded by the papC gene cluster), fimbrial adhesin (sfaS), adhesins from the Afa/Dr family of adhesins (afa/dra), ferri-aerobactin receptor (iutA), and type II capsule synthesis (kpsMT II) genes (Cergole-Novella et al., 2010Cergole-Novella MC, Guth BEC, Castanheira M et al.(2010) First description of bla (CTX-M-14)- and bla (CTX-M-15)-producing Escherichia coliisolates in Brazil. Microb Drug Resist 16:177–184.), and other 18 virulence associated genes cnf (cytotoxic necrotizing factor), cnf1 (cytotoxic necrotizing factor type 1), crl(curli regulator), csgA (curli structural subunit), irp2 (yersiniabactin), fimH (type 1 fimbria), cvaC (colicin V), ibe10 (brain microvascular endothelial cells invasion), traT (serum resistance), iha (adhesin similar to IrgA), espI and espP (extracellular serine proteases), vat(vacuolating cytotoxin), spate (serine protease autotransporters of the Enterobacteriaceae), aggR, aatA, aggA and aafA (molecular markers of EAEC) were assayed by PCR amplification using the primers and amplification conditions described (Blanco et al., 1996Blanco M, Blanco IE, Blanco J et al. (1996) Polymerase chain reaction for detection of Escherichia colistrains producing cytotoxic necrotizing factor type 1 and type 2 (CNFl and CNF2). J Microbiol Methods 26:95–101.; Maurer et al., 1998Maurer JJ, Brown TP, Steffens WL et al. (1998) The occurrence of ambient temperature-regulated adhesins, curli, and the temperature- sensitive hemagglutinin tsh among avian Escherichia coli. Avian Dis 42:106–118.; Schubert et al., 1998Schubert S, Rakin A, Karch H et al. (1998) Prevalence of the “High-Pathogenicity Island” of YersiniaSpecies among Escherichia coli strains that are pathogenic to humans. Infect Immunity 66:480–485.; Johnson and Stell, 2000Johnson JR, Stell AL (2000) Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272.; Tarr et al., 2000Tarr PI, Bilge SS, James C et al. (2000) Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun 68:1400–1407.; Schmidt et al., 2001Schmidt H, Zhang WL, Hemmrich U et al. (2001) Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli Infect Immun 69:6863–6873.; Beutin et al., 2005Beutin L, Kaulfuss S, Herold S et al. (2005) Genetic analysis of Enteropathogenic and Enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and Pulsed-Field Gel Electrophoresis. J Clin Microbiol 43:1552–1563.; Parham et al., 2005Parham NJ, Pollard SJ, Chaudhuri RR et al. (2005) Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli. J Clin Microbiol 43:2425–2434.; Kotlowski et al., 2007Kotlowski R, Bernstein CN, Sepehri S et al. (2007) High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56:669–675.; Monteiro et al., 2009Monteiro BT, Campos LC, Sircili MP et al. (2009) The dispersin-encoding gene (aap) is not restricted to enteroaggregative Escherichia coli. Diag Microbiol Infect Dis 65:81–84.). Phenotypic assays were also carried out for detection of hemolytic activity (Beutin, 1991Beutin L (1991) The different hemolysins of Escherichia coli. Med Microbiol Immunol 180:167–182.) and type 1 fimbriae (Hancock, 2011Hancock V, Witso IL, Klemm P (2011) Biofilm formation as a function of adhesin, growth medium, substratum and strain type. Int J Med Microbiol 301:570–576.). The genes crl-csgA-fimH, traT, irp2, iutA-spates and kpsMTII were identified in 100%, 83.3%, 75%, 50% and 41.7% of the isolates, respectively. Presence of four up to 12 virulence genes was observed among isolates resistant to one or two antimicrobials. Different virulence profiles were identified among the isolates studied (Table 1). The three CTX-M-producing E. coli isolates belonging to O102:H6 and ONT:HNM serotypes harbored 5 of the 20 virulence markers studied. On the other hand, the CTX-M producers albeit carrying resistance to 10 different antimicrobials presented only five of the 18 virulence genes analyzed. These results were different from those reported by Pitout et al. (2012)Pitout JD (2012) Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3:1–7. who found that virulence factors were in general more prevalent among CTX-M producers. Interestingly, albeit controversial, it seems that for some of the isolates studied herein the acquisition of resistance genes was paralleled by a lower number of virulence factors. In addition, it was interesting to observe that the more complex set of virulence markers occur among isolates from urinary tract infections and non-CTX-M producers carrying from two to four antimicrobial resistant genes. A larger number of virulence genes were found among isolates of serogroups O1, O4, and O6 obtained from community infections, some of them harboring cnf1, fimH, papC, and expressing α-Hly (Table 1). Blum et al. (1995)Blum G, Falbo V, Caprioli A et al. (1995) Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett 126:189–195. reported that cnf1, fimH, papC, and hlyA genes are on the pathogenicity island II of the uropathogenic E. coli strain J96 (O4:K6). Members of the serine protease autotransporters of Enterobacteriaceae (SPATEs) family can contribute to the virulence of different E. coli pathotypes (Parham et al., 2005Parham NJ, Pollard SJ, Chaudhuri RR et al. (2005) Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli. J Clin Microbiol 43:2425–2434.). Indeed, 50% of the isolates presently studied carried spate, and vat sequence member of the SPATE family was present in three urosepsis E. coli isolates (Table 1). The invasion of brain endothelium gene ibe10 was carried by two of the 6 urosepsis isolates studied herein (Table 1). Invasion assays, mentioned before, showed detachment of CaCo-2 and HK-2 cell monolayers after 3 h of contact by the E. coli 47 isolate that presented αHly and carried cnf1 and papC genes (Table 1). Earlier studies implicated haemolysin production and P fimbriae as a cause of cell detachment (Marques et al., 1995Marques LR, Abe CM, Griffin PM et al. (1995) Association between alpha-hemolysin production and HeLa cell-detaching activity in fecal isolates of Escherichia coli. J Clin Microbiol 33:2707–2709.). However, Paciorek (2002)Paciorek J (2002) Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J Med Microbiol 51:548–556. observed that cell detachment was not associated with the presence of cnf1 gene.
In conclusion, one can suggest that the combination of biofilm formation, ability to adhere to different epithelial cells, mostly to HK-2 cells, a panel of different virulence markers, and resistance to multiple antimicrobials certainly have contributed to the success of infection caused by these aggregative extraintestinal E. coli isolates.
Acknowledgments
We thank Dr. N O Camara for kindly providing HK-2 human epithelial cells used in the adherence assay. This study was supported by grants from Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq) (to BECG) and Coordenação de Aperfeicoamento de Pessoal de Nível Superior (CAPES). There is no conflict of interest to be disclosed.
References
- Beutin L (1991) The different hemolysins of Escherichia coli Med Microbiol Immunol 180:167–182.
- Beutin L, Kaulfuss S, Herold S et al. (2005) Genetic analysis of Enteropathogenic and Enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and Pulsed-Field Gel Electrophoresis. J Clin Microbiol 43:1552–1563.
- Biscola FT, Abe CM, Guth BEC (2011) Determination of adhesin gene sequences in, and biofilm formation by, O157 and non-O157 Shiga toxin-producing Escherichia coli strains isolated from different sources. Appl Environ Microbiol 77: 2201–2208.
- Blanco M, Blanco IE, Blanco J et al. (1996) Polymerase chain reaction for detection of Escherichia colistrains producing cytotoxic necrotizing factor type 1 and type 2 (CNFl and CNF2). J Microbiol Methods 26:95–101.
- Blum G, Falbo V, Caprioli A et al. (1995) Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol Lett 126:189–195.
- Boll EJ, Struve C, Boisen N et al. (2013) Role of Enteroaggregative Escherichia coli Virulence Factors in Uropathogenesis Infect Immun 81:1164–1171.
- Cergole-Novella MC, Guth BEC, Castanheira M et al.(2010) First description of bla (CTX-M-14)- and bla (CTX-M-15)-producing Escherichia coliisolates in Brazil. Microb Drug Resist 16:177–184.
- Cravioto A, Gross RJ, Scotland SM et al. (1979) An adhesive factor found in strains of E. coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol 3:95–99.
- Hancock V, Witso IL, Klemm P (2011) Biofilm formation as a function of adhesin, growth medium, substratum and strain type. Int J Med Microbiol 301:570–576.
- Johnson JR, Stell AL (2000) Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272.
- Johnson JR, O’Bryan TT, Kuskowski M et al. (2001) Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect Immun 69:5363–5374.
- Kaper JB, Nataro JP, Harry LT et al. (2004) Pathogenic Escherichia coli Nat Rev Microbiol 2:123–140.
- Kotlowski R, Bernstein CN, Sepehri S et al. (2007) High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56:669–675.
- Marques LR, Abe CM, Griffin PM et al. (1995) Association between alpha-hemolysin production and HeLa cell-detaching activity in fecal isolates of Escherichia coli J Clin Microbiol 33:2707–2709.
- Maurer JJ, Brown TP, Steffens WL et al. (1998) The occurrence of ambient temperature-regulated adhesins, curli, and the temperature- sensitive hemagglutinin tsh among avian Escherichia coli Avian Dis 42:106–118.
- Monteiro BT, Campos LC, Sircili MP et al. (2009) The dispersin-encoding gene (aap) is not restricted to enteroaggregative Escherichia coli Diag Microbiol Infect Dis 65:81–84.
- Paciorek J (2002) Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J Med Microbiol 51:548–556.
- Parham NJ, Pollard SJ, Chaudhuri RR et al. (2005) Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli J Clin Microbiol 43:2425–2434.
- Peirano G, Mulvey GL, Armstrong GD et al. (2013) Virulence potential and adherence properties of Escherichia coli that produce CTX-M and NDM β-lactamases. J Med Microbiol 62:525–530.
- Pitout JD (2012) Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3:1–7.
- Robins-Browne RM, Bennett-Wood V (1992) Quantitative assessment of the ability of Escherichia coli to invade cultured animal cells. Microb Pathog 12:159–164.
- Schmidt H, Zhang WL, Hemmrich U et al. (2001) Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli Infect Immun 69:6863–6873.
- Schubert S, Rakin A, Karch H et al. (1998) Prevalence of the “High-Pathogenicity Island” of YersiniaSpecies among Escherichia coli strains that are pathogenic to humans. Infect Immunity 66:480–485.
- Stewart PS (1994) Biofilm Accumulation Model That Predicts Antibiotic Resistance of Pseudomonas aeruginosa Biofilms. Antimicrob Agents Chemother 38:1052–1058.
- Tarr PI, Bilge SS, James C et al. (2000) Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun 68:1400–1407.
- Uhlich GA, Cooke PH, Solomon EB (2006) Analyses of the red-dry rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol 72:2564–2572.
- Zogaj X, Nimtz M, Rohde M et al. (2001) The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463.
Publication Dates
-
Publication in this collection
Jan-Mar 2015
History
-
Received
27 Jan 2014 -
Accepted
06 June 2014