Abstract
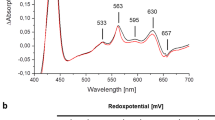
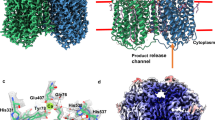
Fumarate reductase couples the reduction of fumarate to succinate to the oxidation of quinol to quinone, in a reaction opposite to that catalysed by the related complex II of the respiratory chain (succinate dehydrogenase). Here we describe the crystal structure at 2.2 Å resolution of the three protein subunits containing fumarate reductase from the anaerobic bacterium Wolinella succinogenes. Subunit A contains the site of fumarate reduction and a covalently bound flavin adenine dinucleotide prosthetic group. Subunit B contains three iron–sulphur centres. The menaquinol-oxidizing subunit C consists of five membrane-spanning, primarily helical segments and binds two haem b molecules. On the basis of the structure, we propose a pathway of electron transfer from the dihaem cytochrome b to the site of fumarate reduction and a mechanism of fumarate reduction. The relative orientations of the soluble and membrane-embedded subunits of succinate:quinone oxidoreductases appear to be unique.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Saraste,M. Oxidative phosphorylation at the fin de siècle. Science 283, 1488–1493 (1999).
Kröger,A. Fumarate as terminal acceptor of phosphorylative electron transport. Biochim. Biophys. Acta 505, 129–145 (1978).
Kröger,A. et al. Bacterial fumarate respiration. Arch. Microbiol. 158, 311–314 (1992).
Hägerhäll,C. Succinate:quinone oxidoreductases. Variations on a conserved theme. Biochim. Biophys. Acta 1320, 107–141 (1997).
Iverson,T. M., Luna-Chavez,C., Cecchini,G. & Rees,D. C. Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284, 1961–1966 (1999).
Lauterbach,F. et al. The fumarate reductase operon of Wolinella succinogenes. Sequence and expression of the frdA and frdB genes. Arch. Microbiol. 154, 386–393 (1990).
Iwata,S., Ostermeier,C., Ludwig,B. & Michel,H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376, 660–669 (1995).
Kuriyan,J. et al. Convergent evolution of similar function in two structurally divergent enzymes. Nature 352, 172–174 (1991).
Holm,L. & Sander,C. Mapping the protein universe. Science 273, 595–602 (1996).
Mattevi,A. et al. Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family. Structure 7, 745–756 (1999).
Singer,T. P. & McIntire,W. S. Covalent attachment of flavin to flavoproteins: Occurrence, assay, and synthesis. Methods Enzymol. 106, 369–378 (1984).
Cole,S. T., Condon,C., Lemire,B. D. & Weiner,J. H. Molecular biology, biochemistry and bioenergetics of fumarate reductase, a complex membrane-bound iron-sulfur flavoenzyme of Escherichia coli. Biochim. Biophys. Acta 811, 381–403 (1985).
Kenny,W. C. & Kröger,A. The covalently bound flavin of Vibrio succinogenes succinate dehydrogenase. FEBS Lett. 73, 239–243 (1977).
van Hellemond,J. J. & Tielens,A. G. M. Expression and functional properties of fumarate reductase. Biochem. J. 304, 321–331 (1994).
Schröder,I. et al. Identification of active site residues of Escherichia coli fumarate reductase by site-directed mutagenesis. J. Biol. Chem. 266, 13572–13579 (1991).
Unden,G. & Kröger,A. An essential sulfhydryl group at the substrate binding site of the fumarate reductase of Vibrio succinogenes. FEBS Lett. 117, 323–326 (1980).
Tomb,J.-F. et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547 (1997).
Flachmann,R. et al. Molecular biology of pyridine nucleotide biosynthesis in Escherichia coli. Cloning and characterization of quinolate synthesis genes nadA and nadB. Eur. J. Biochem. 175, 221–228 (1988).
Vik,S. B. & Hatefi,Y. Possible occurrence and role of an essential histidyl residue in succinate dehydrogenase. Proc. Natl Acad. Sci. USA 78, 6749–6753 (1981).
Tsukihara,T. et al. Structure of the [2Fe-2S] ferredoxin I from Aphantothece sacrum at 2.2 Ångstroms resolution. J. Mol. Biol. 216, 399–410 (1990).
Körtner et al. Wolinella succinogenes fumarate reductase contains a dihaem cytochrome b. Mol. Microbiol. 4, 855–860 (1990).
Hägerhäll,C. & Hederstedt,L. A structural model for the membrane-integral domain of succinate:quinone oxidoreductases. FEBS Lett. 389, 25–31 (1996).
Simon,J. et al. Deletion and site-directed mutagenesis of the Wolinella succinogenes fumarate reductase operon. Eur. J. Biochem. 251, 418–426 (1998).
Xia,D. et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277, 60–66 (1997).
Unden,G., Albracht,S. P. J. & Kröger,A. Redox potentials and kinetic properties of fumarate reductase complex from Vibrio succinogenes. Biochim. Biophys. Acta 767, 460–469 (1984).
Salerno,J. C. Electron transfer in succinate:ubiquinone reductase and quinol:fumarate reductase. Biochem. Soc. Trans. 19, 599–605 (1991).
Lancaster,C. R. D. Quinone-binding sites in membrane proteins: what can we learn from the Rhodopseudomonas viridis reaction centre? Biochem. Soc. Trans. 27, 591–596 (1999).
Hederstedt,L. Bioenergetics—Respiration without O2. Science 284, 1941–1942 (1999).
Bronder,M., Mell,H., Stupperich,E. & Kröger,A. Biosynthetic pathways of Vibrio succinogenes growing with fumarate as terminal electron acceptor and sole carbon source. Arch. Microbiol. 131, 213–223 (1982).
Unden,G., Hackenberg,H. & Kröger,A. Isolation and functional aspects of the fumarate reductase involved in the phosphorylative electron transport of Vibrio succinogenes. Biochim. Biophys. Acta 591, 275–288 (1980).
Tsiotis,G. in A Practical Guide to Membrane Protein Purification (eds von Jagow, G. & Schägger, H.) 149–159 (Academic, San Diego, 1994).
McPherson,A. Crystallization of Biological Macromolecules 304–305 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1999).
Otwinowski,Z. & Minor,W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computation Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
De La Fortelle,E. & Bricogne,G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997).
Otwinowski,Z. in Proc. CCP4 Study Weekend, 25-26 Jan 1991, Isomorphous Replacement and Anomalous Scattering (eds Wolf, W., Evans, P. R. & Leslie, A. G. W.) 80–86 (SERC Daresbury Laboratory, Warrington, UK, 1991).
Cowtan,K. dm: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 30, 34–38 (1994).
Jones,T. A., Zou,J. Y., Cowan,S. W. & Kjeldgaard,M. Improved methods for building protein models in electron density maps and the location of errors within these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger,A. T. et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Brünger,A. T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 (1992).
Kleywegt,G. J. & Brünger,A. T. Checking your imagination: Applications of the free R value. Structure 4, 897–904 (1996).
Luzzati,P. V. Traitement statistique des erreurs dans la détermination des structures cristallines. Acta Crystallogr. 5, 802–810 (1952).
Read,R. J. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42, 140–149 (1986).
Engh,R. H. & Huber,R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47, 392–400 (1991).
Lancaster,C. R. D. & Michel, H. The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB. Structure 5, 1339–1359 (1997).
Kraulis,P. J. MolScript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Esnouf,R. M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graphics Mod. 15, 132–134 (1997).
Esnouf,R. M. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55, 938–940 (1999).
Kabsch,W. & Sander,C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Merritt,E. A. & Bacon,D. J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Acknowledgements
We thank A. Mattevi and co-workers for providing the coordinates of the E. coli L-aspartate oxidase before release for the Protein Data Bank; S. Gemeinhardt, O. Schürmann (both Institut für Mikrobiologie), C. Weiss, B. Marx, C. Münke, C. Wardenberg and D. Vinzenz (all at MPI Biophysik) for technical assistance; A. Thompson (EMBL Grenoble), G. Leonard and E. Mitchell (both ESRF) for support during data acquisition and preliminary data processing at ESRF Grenoble beamline BM14; and L.-O. Essen for providing purified tetrakis-(acetoxymercuri)-methane (TAMM). This work was supported by the Deutsche Forschungsgemeinschaft (SFB 472, grants to A.K. and H.M.) and the Max-Planck-Gesellschaft.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
Supplementary Information (PDF 58 kb)
Rights and permissions
About this article
Cite this article
Lancaster, C., Kröger, A., Auer, M. et al. Structure of fumarate reductase from Wolinella succinogenes at 2.2 Å resolution. Nature 402, 377–385 (1999). https://doi.org/10.1038/46483
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/46483
This article is cited by
-
A signaling complex of adenylate cyclase CyaC of Sinorhizobium meliloti with cAMP and the transcriptional regulators Clr and CycR
BMC Microbiology (2023)
-
Molecular basis of maintaining an oxidizing environment under anaerobiosis by soluble fumarate reductase
Nature Communications (2018)
-
Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas
Scientific Reports (2018)
-
Multiplicity and specificity of siderophore uptake in the cyanobacterium Anabaena sp. PCC 7120
Plant Molecular Biology (2016)
-
MetalS3, a database-mining tool for the identification of structurally similar metal sites
JBIC Journal of Biological Inorganic Chemistry (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.