Abstract
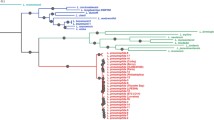
The order Legionellales contains several clinically important microorganisms. Although members of this order are well-studied for their pathogenesis, there is a paucity of reliable characteristics distinguishing members of this order and its constituent genera. Genome sequences are now available for 73 Legionellales species encompassing ≈90% of known members from different genera. With the aim of understanding evolutionary relationships and identifying reliable molecular characteristics that are specific for this order and its constituent genera, detailed phylogenetic and comparative analyses were conducted on the protein sequences from these genomes. A phylogenomic tree was constructed based on 393 single copy proteins that are commonly shared by the members of this order to delineate the evolutionary relationships among its members. In parallel, comparative analyses were performed on protein sequences from Legionellales genomes to identify novel molecular markers consisting of conserved signature indels (CSIs) that are specific for different clades and genera. In the phylogenomic tree and in an amino acid identity matrix based on core proteins, members of the genera Aquicella, Coxiella, Legionella and Rickettsiella formed distinct clades confirming their monophyly. In these studies, Diplorickettsia massiliensis exhibited a close relationship to members of the genus Rickettsiella. The results of our comparative genomic analyses have identified 59 highly specific molecular markers consisting of CSIs in diverse proteins that are uniquely shared by different members of this order. Four of these CSIs are specific for all Legionellales species, except the two deeper-branching “Candidatus Berkiella” species, providing means for identifying members of this order in molecular terms. Twenty four, 7 and 6 CSIs are uniquely shared by members of the genera Legionella, Coxiella and Aquicella, respectively, identifying these groups in molecular terms. The descriptions of these three genera are emended to include information for their novel molecular characteristics. We also describe 12 CSIs that are uniquely shared by D. massiliensis and different members of the genus Rickettsiella. Based on these results, we are proposing an integration of the genus Diplorickettsia with Rickettsiella. Three other CSIs suggest that members of the genera Coxiella and Rickettsiella shared a common ancestor exclusive of other Legionellales. The described molecular markers, due to their exclusivity for the indicated taxa/genera, provide important means for the identification of these clinically important microorganisms and for discovering novel properties unique to them.








Similar content being viewed by others
Data availability
All of the genomes sequences analyzed in this study are publicly available. The accession numbers of all protein sequences used are shown in different Figures and Tables and detailed information for them is provided in Supplemental attachment.
References
Adeolu M, Alnajar S, Naushad S, Gupta S (2016) Genome-based phylogeny and taxonomy of the “Enterobacteriales”: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599
Ahmod NZ, Gupta RS, Shah HN (2011) Identification of a Bacillus anthracis specific indel in the yeaC gene and development of a rapid pyrosequencing assay for distinguishing B. anthracis from the B. cereus group. J Microbiol Methods 87:278–285
Albuquerque L, Rainey FA, Da Costa MS (2020) Aquicella. In: Whitman WB, DeVos P, Dedysh S, Hedlund B, Kampfer P, Rainey F, Trujillo ME, Bowman JP, Brown DR, Glockner F, Oren A, Paster BJ, Wade W, Ward N, Busse H-J, Reysenbach A-L (eds) Bergey’s manual of systematics of archaea and bacteria, online. Wiley, Hoboken
Baldauf SL (2003) Phylogeny for the faint of heart: a tutorial. Trends Genet 19:345–351
Bhandari V, Gupta RS (2014) Molecular signatures for the phylum (class) Thermotogae and a proposal for its division into three orders (Thermotogales, Kosmotogales ord. nov. and Petrotogales ord. nov.) containing four families (Thermotogaceae, Fervidobacteriaceae fam. nov., Kosmotogaceae fam. nov. and Petrotogaceae fam. nov.) and a new genus Pseudothermotoga gen. nov. with five new combinations. Antonie Van Leeuwenhoek 105:143–168
Brenner DJ, Steigerwalt AG, McDade JE (1979) Classification of the Legionnaires’ disease bacterium: Legionella pneumophila, genus novum, species nova of the family Legionellceae, familia nova. Ann Intern Med 90:656–658
Brown A, Garrity GM, Vickers RM (1981) Fluoribacter dumoffii (Brenner et al.) comb. nov. and Fluoribacter gormanii (Morris et al.) comb. nov. Int J Syst Evol Microbiol 31:111–115
Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O et al (2016) Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet 48:167–175
Buysse M, Duron O (2020) Two novel Rickettsia species of soft ticks in North Africa: 'Candidatus Rickettsia africaseptentrionalis’ and ‘Candidatus Rickettsia mauretanica’. Ticks Tick Borne Dis 11:101376
Buysse M, Plantard O, McCoy KD, Duron O, Menard C (2019) Tissue localization of Coxiella-like endosymbionts in three European tick species through fluorescence in situ hybridization. Ticks Tick Borne Dis 10:798–804
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T (2009) TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Cianciotto NP, Hilbi H, Buchrieser C (2013) Legionnaires’ disease. In: Rosenberg E, Delong E, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: human microbiology. Springer, Berlin, pp 147–217
Cordevant C, Tang JS, Cleland D, Lange M (2003) Characterization of members of the Legionellaceae family by automated ribotyping. J Clin Microbiol 41:34–43
Cutino-Jimenez AM, Menck CFM, Cambas YT, Diaz-Perez JC (2020) Protein signatures to identify the different genera within the Xanthomonadaceae family. Braz J Microbiol 51:1515–1526
Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A et al (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39:W13–W17
Dobritsa AP, Samadpour M (2019) Reclassification of Burkholderia insecticola as Caballeronia insecticola comb. nov. and reliability of conserved signature indels as molecular synapomorphies. Int J Syst Evol Microbiol 69:2057–2063
Duron O, Jourdain E, McCoy KD (2014) Diversity and global distribution of the Coxiella intracellular bacterium in seabird ticks. Ticks Tick Borne Dis 5:557–563
Duron O, Doublet P, Vavre F, Bouchon D (2018) The importance of revisiting legionellales diversity. Trends Parasitol 34:1027–1037
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195
Felsenstein J (2004) Inferring phylogenies. Sinauer Associates Inc, Sunderland
Fields BS, Benson RF, Besser RE (2002) Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev 15:506–526
Fournier PE, Raoult D (2015) Rickettsiella. In: Whitman WB, DeVos P, Dedysh S, Hedlund B, Kampfer P, Rainey F, Trujillo ME, Bowman JP, Brown DR, Glockner F, Oren A, Paster BJ, Wade W, Ward N, Busse H-J, Reysenbach A-L (eds) Bergey’s manual of systematics of archaea and bacteria, online. Wiley, Hoboken
Frutos R, Federici BA, Revet B, Bergoin M (1994) Taxonomic studies of Rickettsiella, Rickettsia, and Chlamydia using genomic DNA. J Invertebr Pathol 63:294–300
Fry NK, Warwick S, Saunders NA, Embley TM (1991) The use of 16S ribosomal RNA analyses to investigate the phylogeny of the family Legionellaceae. J Gen Microbiol 137:1215–1222
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152
Gao B, Gupta RS (2005) Conserved indels in protein sequences that are characteristic of the phylum Actinobacteria. Int J Syst Evol Microbiol 55:2401–2412
Garrity GM, Brown A, Vickers RM (1980) Tatlockia and Fluoribacter: two new genera of organisms resembling Legionella pneumophila. Int J Syst Evol Microbiol 30:609–614
Garrity GM, Bell J, Lilburn T (2005) Legionellales ord. nov. In: Brenner D, Krieg N, Staley J, Garrity GM, Boone D, De Vos P, Goodfellow M, Rainey F, Schleifer KH (eds) Bergey’s manual of systematic bacteriology part B gammaproteobacteria. Springer, Boston, pp 210–247
Graells T, Ishak H, Larsson M, Guy L (2018) The all-intracellular order Legionellales is unexpectedly diverse, globally distributed and lowly abundant. FEMS Microbiol Ecol 94:fiy185
Grattard F, Ginevra C, Riffard S, Ros A, Jarraud S et al (2006) Analysis of the genetic diversity of Legionella by sequencing the 23S–5S ribosomal intergenic spacer region: from phylogeny to direct identification of isolates at the species level from clinical specimens. Microbes Infect 8:73–83
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W et al (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Gupta RS (1998) Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev 62:1435–1491
Gupta RS (2014) Identification of conserved indels that are useful for classification and evolutionary studies. In: Methods in microbiology. Oxford/Elsevier, pp 153–182
Gupta RS (2016) Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin’s views on classification. FEMS Microbiol Rev 40:520–553
Gupta RS, Lo B, Son J (2018) Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol 9:67
Gupta RS, Patel S, Saini N, Chen S (2020) Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int J Syst Evol Microbiol 70:5753–5798
Hamilton KA, Prussin AJI, Ahmed W, Hass CN (2018) Outbreaks of Legionnaires’ disease and Pontiac Fever 2006–2017. Curr Environ Health Rep 5:263–271
Hasegawa M, Kishino H, Saitou N (1991) On the maximum likelihood method in molecular phylogenetics. J Mol Evol 32:443–445
Hébert GA, Steigerwalt AG, Brenner DJ (1980) Legionella micdadei in validation list no. 5. Int J Syst Evol Microbiol 90:676–677
Herbeck JT, Degnan PH, Wernegreen JJ (2005) Nonhomogeneous model of sequence evolution indicates independent origins of primary endosymbionts within the enterobacteriales (gamma-Proteobacteria). Mol Biol Evol 22:520–532
Hookey JV, Birtles RJ, Saunders NA (1995) Intergenic 16S rRNA gene (rDNA)-23S rDNA sequence length polymorphisms in members of the family Legionellaceae. J Clin Microbiol 33:2377–2381
Hookey JV, Saunders NA, Fry NK, Birtles RJ, Harrison TG (1996) Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like Ameobal Pathogens. Int J Syst Evol Microbiol 46:526–531
Horn M, Wagner M (2004) Bacterial endosymbionts of free-living amoebae. J Eukaryot Microbiol 51:509–514
Hugoson E, Ammunet T, Guy L (2020) Host-adaptation in Legionellales is 2.4 Ga, coincident with eukaryogenesis. BioRxiv. https://doi.org/10.1101/852004
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal x. Trends Biochem Sci 23:403–405
Jiang L, Wang D, Kim J-S, Lee JH, Kim D-H et al (2021) Reclassification of genus Izhakiella into the family Erwiniaceae based on phylogenetic and genomic analyses. Int J Syst Evol Microbiol 70:3541–3546
Keeling PJ, Palmer JD (2000) Parabasalian flagellates are ancient eukaryotes. Nature 405:635–637
Khadka B, Gupta RS (2017) Identification of a conserved 8 aa insert in the PIP5K protein in the Saccharomycetaceae family of fungi and the molecular dynamics simulations and structural analysis to investigate its potential functional role. Proteins 85:1454–1467
Khodr A, Kay E, Gomez-Valero L, Ginevra C, Doublet P et al (2016) Molecular epidemiology, phylogeny and evolution of Legionella. Infect Genet Evol 43:108–122
Ko KS, Lee HK, Park MY, Lee KH, Yun YJ et al (2002) Application of RNA polymerase beta-subunit gene (rpoB) sequences for the molecular differentiation of Legionella species. J Clin Microbiol 40:2653–2658
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Lory S (2014b) The family Legionellaceae. In: Rosenberg E, Delong E, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: gammaproteobacteria. Springer, Berlin, pp 387–389
Lory S (2014a) The family Coxiellaceae. In: Rosenberg E, Delong E, Lory S, Stackebrandt E, Tsao HF (eds) The prokaryotes: gammaproteobacteria. Springer, Berlin, pp 197–198
Mathew MJ, Subramanian G, Nguyen TT, Robert C, Mediannikov O et al (2012) Genome sequence of Diplorickettsia massiliensis, an emerging Ixodes ricinus-associated human pathogen. J Bacteriol 194:3287
Mediannikov O, Sekeyova Z, Birg ML, Raoult D (2010) A novel obligate intracellular gamma-proteobacterium associated with ixodid ticks, Diplorickettsia massiliensis, Gen. Nov., Sp. Nov. PLoS ONE 5:e11478
Mehari YT, Jason HB, Redding KS, Mariappan PVG, Gunderson JH et al (2016) Description of 'Candidatus Berkiella aquae’ and 'Candidatus Berkiella cookevillensis’, two intranuclear bacteria of freshwater amoebae. Int J Syst Evol Microbiol 66:536–541
Moreira D, Philippe H (2000) Molecular phylogeny: pitfalls and progress. Int Microbiol 3:9–16
Naushad HS, Lee B, Gupta RS (2014) Conserved signature indels and signature proteins as novel tools for understanding microbial phylogeny and systematics: identification of molecular signatures that are specific for the phytopathogenic genera Dickeya, Pectobacterium and Brenneria. Int J Syst Evol Microbiol 64:366–383
Nooroong P, Trinachartvanit W, Baimai V, Ahantarig A (2018) Phylogenetic studies of bacteria (Rickettsia, Coxiella, and Anaplasma) in Amblyomma and Dermacentor ticks in Thailand and their co-infection. Ticks Tick Borne Dis 9:963–971
Notredame C, Higgins DG, Heringa J (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217
Parte AC (2018) LPSN—the list of prokaryotic names with standing in nomenclature. Int J Syst Evol Microbiol 68:1825–1829
Philip CB (1948) Comments on the name of the Q-fever organism. Public Health Report 1948; 63:58. Public Health Report 63:58.
Philip CB (1956) Comments on the classification of the order Rickettsiales. Can J Microbiol 2:261–270
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490
Ratcliff RM (2013) Sequence-based identification of legionella. Methods Mol Biol 954:57–72
Rivera MC, Lake JA (1992) Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257:74–76
Rokas A, Holland PW (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15:454–459
Rubin CJ, Thollesson M, Kirsebom LA, Herrmann B (2005) Phylogenetic relationships and species differentiation of 39 Legionella species by sequence determination of the RNase P RNA gene rnpB. Int J Syst Evol Microbiol 55:2039–2049
Santos P, Pinhal I, Rainey FA, Empadinhas N, Costa J et al (2003) Gamma-proteobacteria Aquicella lusitana gen. nov., sp. nov., and Aquicella siphonis sp. nov. infect protozoa and require activated charcoal for growth in laboratory media. Appl Environ Microbiol 69:6533–6540
Schulz F, Tyml T, Pizzetti I, Dykova I, Fazi S et al (2015) Marine amoebae with cytoplasmic and perinuclear symbionts deeply branching in the Gammaproteobacteria. Sci Rep 5:13381
Shaw EI, Voth DE (2019) Coxiella burnetii: a pathogenic intracellular acidophile. Microbiology 165:1–3
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Singh B, Gupta RS (2009) Conserved inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins are essential for cellular growth. Mol Genet Genom 281:361–373
Skerman VBD, McGowan V, Sneath PHA (1980) Approved lists of bacterial names. Int J Syst Bacteriol 30:225–420
Springer MS, Stanhope MJ, Madsen O, de Jong WW (2004) Molecules consolidate the placental mammal tree. Trends Ecol Evol 19:430–438
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Subramanian G, Mediannikov O, Angelakis E, Socolovschi C, Kaplanski G et al (2012) Diplorickettsia massiliensis as a human pathogen. Eur J Clin Microbiol Infect Dis 31:365–369
Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E et al (2013) Microbial genomic taxonomy. BMC Genom 14:913
Tindall BJ (2020) Taking a Closer Look at the Valid Publication and Authorship of Legionella bozemanae Brenner et al. 1980, Fluoribacter bozemanae Garrity et al. 1980, Legionella pittsburghensis Pasculle et al. 1980, Legionella micdadei Hebert et al. 1980 and Tatlockia micdadei (Hebert et al. 1980) Garrity et al. 1980. Curr Microbiol 77:146–153
Tsao HF, Scheikl U, Volland JM, Kohsler M, Bright M et al (2017) “Candidatus Cochliophilus cryoturris” (Coxiellaceae), a symbiont of the testate amoeba Cochliopodium minus. Sci Rep 7:3394
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699
Winn WC (2015) Legionella. In: Whitman WB, DeVos P, Dedysh S, Hedlund B, Kampfer P, Rainey F, Trujillo ME, Bowman JP, Brown DR, Glockner F, Oren A, Paster BJ, Wade W, Ward N, Busse H-J, Reysenbach A-L (eds) Bergey’s Manual of systematics of archaea and bacteria, online. Wiley, Hoboken, pp 1–44
Wong SY, Paschos A, Gupta RS, Schellhorn HE (2014) Insertion/deletion-based approach for the detection of Escherichia coli O157:H7 in freshwater environments. Environ Sci Technol 48:11462–11470
Yarza P, Richter M, Peplies J, Euzeby J, Amann R et al (2008) The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31:241–250
Zhan XY, Hu CH, Zhu QY (2016) Targeting single-nucleotide polymorphisms in the 16S rRNA gene to detect and differentiate Legionella pneumophila and non-Legionella pneumophila species. Arch Microbiol 198:591–594
Funding
This work was supported by the research grant No. 20011106 from the Natural Science and Engineering Research Council of Canada awarded to Dr. Radhey S. Gupta.
Author information
Authors and Affiliations
Contributions
NS was responsible for identification of CSIs, checking their specificity, construction of phylogenetic trees, formatting of figures and writing a draft of the manuscript. RSG obtained the funding for this work, conceptualized and supervised the entire study, checked the specificities and evolutionary significance of the identified CSIs and wrote large part of this manuscript and finalized it. All of the authors have read and approved this manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saini, N., Gupta, R.S. A robust phylogenetic framework for members of the order Legionellales and its main genera (Legionella, Aquicella, Coxiella and Rickettsiella) based on phylogenomic analyses and identification of molecular markers demarcating different clades. Antonie van Leeuwenhoek 114, 957–982 (2021). https://doi.org/10.1007/s10482-021-01569-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01569-9