Abstract
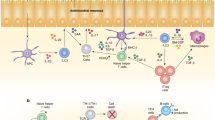
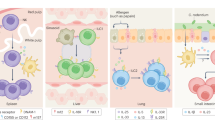
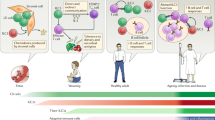
Innate lymphocytes are a diverse population of cells that carry out specialized functions in steady-state homeostasis and during immune challenge. While circulating cytotoxic natural killer (NK) cells have been studied for decades, tissue-resident innate lymphoid cells (ILCs) have only been characterized and studied over the past few years. As ILCs have been largely viewed in the context of helper T-cell biology, models of ILC lineage and function have been founded within this perspective. Notably, tissue-resident innate lymphocytes with cytotoxic potential have been described in an array of tissues, yet whether they are derived from the NK or ILC lineage is only beginning to be elucidated. In this review, we aim to shed light on the identities of innate lymphocytes through the lenses of cell lineage, localization, and timing of differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Nurieva, R. I. et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29, 138–149 (2008).
Vinuesa, C. G., Linterman, M. A., Yu, D. & MacLennan, I. C. Follicular helper T Cells. Annu. Rev. Immunol. 34, 335–368 (2016).
Gerlach, C. et al. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45, 1270–1284 (2016).
Godfrey, D. I., Uldrich, A. P., McCluskey, J., Rossjohn, J. & Moody, D. B. The burgeoning family of unconventional T cells. Nat. Immunol. 16, 1114–1123 (2015).
Godfrey, D. I., Stankovic, S. & Baxter, A. G. Raising the NKT cell family. Nat. Immunol. 11, 197–206 (2010).
Zhou, D. et al. Lysosomal glycosphingolipid recognition by NKT cells. Science 306, 1786–1789 (2004).
Kronenberg, M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu. Rev. Immunol. 23, 877–900 (2005).
Cheroutre, H., Lambolez, F. & Mucida, D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 11, 445–456 (2011).
Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018).
Gury-BenAri, M. et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–46 e13 (2016).
Robinette, M. L. et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16, 306–317 (2015).
Yu X., et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife 3 (2014).
Yang, Q. et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 16, 1044–1050 (2015).
Serafini, N., Vosshenrich, C. A. & Di Santo, J. P. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol. 15, 415–428 (2015).
Klose, C. S. N. et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014).
Constantinides, M. G., McDonald, B. D., Verhoef, P. A. & Bendelac, A. A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014).
Seillet, C. et al. Deciphering the innate lymphoid cell transcriptional program. Cell Rep. 17, 436–447 (2016).
Yu, J., Freud, A. G. & Caligiuri, M. A. Location and cellular stages of natural killer cell development. Trends Immunol. 34, 573–582 (2013).
Fathman, J. W. et al. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood 118, 5439–5447 (2011).
Constantinides, M. G. et al. PLZF expression maps the early stages of ILC1 lineage development. Proc. Natl. Acad. Sci. USA 112, 5123–5128 (2015).
Spits, H. et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat. Rev. Immunol. 13, 145–149 (2013).
Daussy, C. et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211, 563–577 (2014).
Gasteiger, G., Fan, X., Dikiy, S., Lee, S. Y. & Rudensky, A. Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 (2015).
Gordon, S. M. et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 (2012).
Pikovskaya, O. et al. Cutting edge: eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol. 196, 1449–1454 (2016).
Sun, J. C. & Lanier, L. L. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat. Rev. Immunol. 11, 645–657 (2011).
Wong, S. H. et al. Transcription factor RORalpha is critical for nuocyte development. Nat. Immunol. 13, 229–236 (2012).
Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature 517, 293–301 (2015).
Moro, K. et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 463, 540–544 (2010).
Neill, D. R. et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010).
Mjosberg, J. et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity 37, 649–659 (2012).
Zhu, J. Mysterious ILC2 tissue adaptation. Nat. Immunol. 19, 1042–1044 (2018).
Satoh-Takayama, N. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008).
Sanos, S. L. et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+cells. Nat. Immunol. 10, 83–91 (2009).
Luci, C. et al. Influence of the transcription factor RORgammat on the development of NKp46+cell populations in gut and skin. Nat. Immunol. 10, 75–82 (2009).
Buonocore, S. et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375 (2010).
Klose, C. S. & Artis, D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17, 765–774 (2016).
Song, C. et al. Unique and redundant functions of NKp46+ILC3s in models of intestinal inflammation. J. Exp. Med. 212, 1869–1882 (2015).
Eberl, G. et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5, 64–73 (2004).
Lane, P. J. et al. Lymphoid tissue inducer cells: bridges between the ancient innate and the modern adaptive immune systems. Mucosal Immunol. 2, 472–477 (2009).
Sojka, D. K. et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659 (2014).
Takeda, K. et al. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 105, 2082–2089 (2005).
Peng, H. & Tian, Z. Diversity of tissue-resident NK cells. Semin. Immunol. 31, 3–10 (2017).
Yagi, R. et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity 40, 378–388 (2014).
Erick, T. K., Anderson, C. K., Reilly, E. C., Wands, J. R. & Brossay, L. NFIL3 expression distinguishes tissue-resident NK cells and conventional NK-like cells in the mouse submandibular glands. J. Immunol. 197, 2485–2491 (2016).
Juelke, K. & Romagnani, C. Differentiation of human innate lymphoid cells (ILCs). Curr. Opin. Immunol. 38, 75–85 (2016).
Bando, J. K., Liang, H. E. & Locksley, R. M. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat. Immunol. 16, 153–160 (2015).
Mikkola, H. K. & Orkin, S. H. The journey of developing hematopoietic stem cells. Development 133, 3733–3744 (2006).
Kim, S. et al. In vivo developmental stages in murine natural killer cell maturation. Nat. Immunol. 3, 523–528 (2002).
Peng, H. et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Invest. 123, 1444–1456 (2013).
Hydes, T. et al. IL-12 and IL-15 induce the expression of CXCR6 and CD49a on peripheral natural killer cells. Immun. Inflamm. Dis. 6, 34–46 (2018).
Cortez, V. S. et al. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-beta signaling. Nat. Immunol. 18, 995–1003 (2017).
Gao, Y. et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18, 1004–1015 (2017).
Moro, K. et al. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 17, 76–86 (2016).
Ricardo-Gonzalez, R. R. et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat. Immunol. 19, 1093–1099 (2018).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33–41 e7 (2018).
Lim, A. I. et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell 168, 1086–100 e10 (2017).
Lim, A. I. & Di Santo, J. P. ILC-poiesis: ensuring tissue ILC differentiation at the right place and time. Eur. J. Immunol. 49, 11–18 (2018).
Cherrier, D. E., Serafini, N. & Di Santo, J. P. Innate lymphoid cell development: a T cell perspective. Immunity 48, 1091–1103 (2018).
Huang, Y. et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119 (2018).
Kotas, M. E. & Locksley, R. M. Why innate lymphoid cells? Immunity 48, 1081–1090 (2018).
Diefenbach, A. Innate lymphoid cells in the defense against infections. Eur. J. Microbiol Immunol. (Bp) 3, 143–151 (2013).
Fuchs, A. ILC1s in tissue inflammation and infection. Front. Immunol. 7, 104 (2016).
Abt, M. C. et al. Innate immune defenses mediated by two ILC subsets are critical for protection against acute clostridium difficile infection. Cell Host. Microbe 18, 27–37 (2015).
Weizman, O. E. et al. ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808 (2017).
Glasner, A. et al. Recognition and prevention of tumor metastasis by the NK receptor NKp46/NCR1. J. Immunol. 188, 2509–2515 (2012).
Smyth, M. J., Crowe, N. Y. & Godfrey, D. I. NK cells and NKT cells collaborate in host protection from methylcholanthrene-induced fibrosarcoma. Int. Immunol. 13, 459–463 (2001).
Halfteck, G. G. et al. Enhanced in vivo growth of lymphoma tumors in the absence of the NK-activating receptor NKp46/NCR1. J. Immunol. 182, 2221–2230 (2009).
Dadi, S. et al. Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell 164, 365–377 (2016).
Vosshenrich, C. A. et al. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J. Immunol. 174, 1213–1221 (2005).
Ranson, T. et al. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood 101, 4887–4893 (2003).
Robinette, M. L. et al. IL-15 sustains IL-7R-independent ILC2 and ILC3 development. Nat. Commun. 8, 14601 (2017).
Cortez, V. S., Fuchs, A., Cella, M., Gilfillan, S. & Colonna, M. Cutting edge: salivary gland NK cells develop independently of Nfil3 in steady-state. J. Immunol. 192, 4487–4491 (2014).
Satoh-Takayama, N. et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+cell subsets from Id2-dependent precursors. J. Exp. Med. 207, 273–280 (2010).
Burkett, P. R. et al. Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+T cell homeostasis. J. Exp. Med. 200, 825–834 (2004).
Mackay, L. K. et al. The developmental pathway for CD103(+)CD8+tissue-resident memory T cells of skin. Nat. Immunol. 14, 1294–1301 (2013).
Ma, L. J., Acero, L. F., Zal, T. & Schluns, K. S. Trans-presentation of IL-15 by intestinal epithelial cells drives development of CD8alphaalpha IELs. J. Immunol. 183, 1044–1054 (2009).
Lai, Y. G. et al. IL-15 does not affect IEL development in the thymus but regulates homeostasis of putative precursors and mature CD8 alpha alpha+IELs in the intestine. J. Immunol. 180, 3757–3765 (2008).
Klose, C. S. et al. The transcription factor T-bet is induced by IL-15 and thymic agonist selection and controls CD8alphaalpha(+) intraepithelial lymphocyte development. Immunity 41, 230–243 (2014).
Cortez, V. S. et al. Transforming growth factor-beta signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity 44, 1127–1139 (2016).
Mackay, L. K. et al. T-box transcription factors combine with the cytokines TGF-beta and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111 (2015).
Intlekofer, A. M. et al. Effector and memory CD8+T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6, 1236–1244 (2005).
Pegram, H. J., Andrews, D. M., Smyth, M. J., Darcy, P. K. & Kershaw, M. H. Activating and inhibitory receptors of natural killer cells. Immunol. Cell Biol. 89, 216–224 (2011).
Lanier, L. L. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9, 495–502 (2008).
Yokoyama, W. M. & Kim, S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol. Rev. 214, 143–154 (2006).
Gleimer, M. & Parham, P. Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity 19, 469–477 (2003).
Bauer, S. et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729 (1999).
Jamieson, A. M. et al. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity 17, 19–29 (2002).
Savage, P. A. et al. Recognition of a ubiquitous self antigen by prostate cancer-infiltrating CD8 + T lymphocytes. Science 319, 215–220 (2008).
Raulet, D. H. & Guerra, N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat. Rev. Immunol. 9, 568–580 (2009).
Smyth, M. J. et al. NKG2D function protects the host from tumor initiation. J. Exp. Med. 202, 583–588 (2005).
Bando, J. K. & Colonna, M. Innate lymphoid cell function in the context of adaptive immunity. Nat. Immunol. 17, 783–789 (2016).
Acknowledgements
We thank Chun Chou, Briana Nixon, and Efstathios Stamatiades of the Li lab for helpful discussions and critical reading of the manuscript. Work in the Li laboratory was supported by NIAID (R01 CA198280–01 to M.O.L), HHMI (Faculty Scholar Award to M.O.L.), the Ludwig Center for Cancer Immunology (M.O.L.), and the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kansler, E.R., Li, M.O. Innate lymphocytes—lineage, localization and timing of differentiation. Cell Mol Immunol 16, 627–633 (2019). https://doi.org/10.1038/s41423-019-0211-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-019-0211-7
This article is cited by
-
Reversing immunosuppression in the tumor microenvironment of fibrolamellar carcinoma via PD-1 and IL-10 blockade
Scientific Reports (2024)
-
The secretome of Staphylococcus aureus strains with opposite within-herd epidemiological behavior affects bovine mononuclear cell response
Veterinary Research (2023)
-
Chimeric antigen receptor- and natural killer cell receptor-engineered innate killer cells in cancer immunotherapy
Cellular & Molecular Immunology (2021)