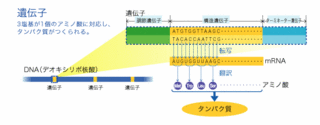
遺伝子
転写単位
遺伝子
遺伝子
遺伝子
【概要】 細胞を工場に例えると、核は本社のコンピュータ室、染色体はハードディスク、遺伝子は特定のプログラムやデータにあたる。つまり遺伝子は一つ一つの蛋白質の構造を決める設計図である。物質としてはDNAとRNAの2種類がある。ウイルスは細胞質や核を持たず、遺伝子を生きた細胞の中に持ち込む。まるでフロッピイディスクのような存在である。
【詳しく】 人間の遺伝子は、23対ある染色体の特定の位置に存在している。これを染色体地図という。遺伝子は信号の始まりの部分から終わりの部分があり、その中間に実際に蛋白質をコードしているエクソン部分と間の無信号域のイントロン部分がある。遺伝子DNAが転写されてRNAができ、さらにイントロン部分が切り出されて、メッセンジャーRNA(mRNA)になる。mRNAは核膜の穴から細胞質に出て蛋白合成に向かう。
遺伝子
GENE
出典: フリー百科事典『ウィキペディア(Wikipedia)』 (2019/11/08 06:58 UTC 版)
『GENE』(ゲーン)は、五百香ノエルによるボーイズラブ小説作品。およびそれを原作としたドラマCD。挿絵は金ひかる。
- 1 GENEとは
- 2 GENEの概要
遺伝子
(Gene から転送)
出典: フリー百科事典『ウィキペディア(Wikipedia)』 (2024/04/12 23:56 UTC 版)
生物学において、遺伝子(いでんし、英: gene、ギリシア語: γένος)という言葉には2つの意味がある。メンデル遺伝子は、遺伝の基本単位である。分子遺伝子は、DNA内のヌクレオチド配列であり、転写されて機能的なRNAを生成する。この分子遺伝子にはタンパク質コード遺伝子と非コード遺伝子の2種類がある[1][2][3][4]。
- ^ a b c Orgogozo V, Peluffo AE, Morizot B (2016). “The "Mendelian Gene" and the "Molecular Gene": Two Relevant Concepts of Genetic Units”. Current Topics in Developmental Biology 119: 1–26. doi:10.1016/bs.ctdb.2016.03.002. PMID 27282022.
- ^ “What is a gene?: MedlinePlus Genetics”. MedlinePlus (2020年9月17日). 2021年1月4日閲覧。
- ^ Hirsch ED (2002). The new dictionary of cultural literacy. Boston: Houghton Mifflin. ISBN 0-618-22647-8. OCLC 50166721
- ^ “Studying Genes”. nigms.nih.gov. 2021年1月15日閲覧。
- ^ Elston RC, Satagopan JM, Sun S (2012). “Genetic terminology”. Statistical Human Genetics. Methods in Molecular Biology. 850. Humana Press. pp. 1–9. doi:10.1007/978-1-61779-555-8_1. ISBN 978-1-61779-554-1. PMC 4450815. PMID 22307690
- ^ a b Johannsen W (1909) (German). Elemente der exakten Erblichkeitslehre [Elements of the exact theory of heredity]. Jena, Germany: Gustav Fischer. p. 124 From p. 124: "Dieses "etwas" in den Gameten bezw. in der Zygote, … – kurz, was wir eben Gene nennen wollen – bedingt sind." (This "something" in the gametes or in the zygote, which has crucial importance for the character of the organism, is usually called by the quite ambiguous term Anlagen [primordium, from the German word Anlage for "plan, arrangement ; rough sketch"]. Many other terms have been suggested, mostly unfortunately in closer connection with certain hypothetical opinions. The word "pangene", which was introduced by Darwin, is perhaps used most frequently in place of Anlagen. However, the word "pangene" was not well chosen, as it is a compound word containing the roots pan (the neuter form of Πας all, every) and gen (from γί-γ(ε)ν-ομαι, to become). Only the meaning of this latter [i.e., gen] comes into consideration here ; just the basic idea – [namely,] that a trait in the developing organism can be determined or is influenced by "something" in the gametes – should find expression. No hypothesis about the nature of this "something" should be postulated or supported by it. For that reason it seems simplest to use in isolation the last syllable gen from Darwin's well-known word, which alone is of interest to us, in order to replace, with it, the poor, ambiguous word Anlage. Thus we will say simply "gene" and "genes" for "pangene" and "pangenes". The word gene is completely free of any hypothesis ; it expresses only the established fact that in any case many traits of the organism are determined by specific, separable, and thus independent "conditions", "foundations", "plans" – in short, precisely what we want to call genes.)
- ^ “1909: The Word Gene Coined”. genome.gov. 2021年3月8日閲覧。 "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..."
- ^ Roth SC (July 2019). “What is genomic medicine?”. Journal of the Medical Library Association (University Library System, University of Pittsburgh) 107 (3): 442–448. doi:10.5195/jmla.2019.604. PMC 6579593. PMID 31258451.
- ^ a b c d Kampourakis K (2017). Making Sense of Genes. Cambridge, UK: Cambridge University Press
- ^ Gericke N, Hagberg M (5 December 2006). “Definition of historical models of gene function and their relation to students' understanding of genetics”. Science & Education 16 (7–8): 849–881. Bibcode: 2007Sc&Ed..16..849G. doi:10.1007/s11191-006-9064-4.
- ^ Meunier R (2022年). “Stanford Encyclopedia of Philosophy: Gene”. Stanford Encyclopedia of Philosophy. 2023年2月28日閲覧。
- ^ Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC (April 2014). “Defining functional DNA elements in the human genome”. Proceedings of the National Academy of Sciences of the United States of America 111 (17): 6131–8. Bibcode: 2014PNAS..111.6131K. doi:10.1073/pnas.1318948111. PMC 4035993. PMID 24753594.
- ^ a b ドーキンス,リチャード 著、日高敏隆/岸由二/羽田節子/垂水雄二 訳『利己的な遺伝子 40周年記念版』紀伊國屋書店、2018年2月15日。ISBN 978-4314011532。
- ^ Stoltz K, Griffiths P (2004). “Genes: Philosophical Analyses Put to the Test”. History and Philosophy of the Life Sciences 26 (1): 5–28. doi:10.1080/03919710412331341621. JSTOR 23333378. PMID 15791804.
- ^ Beadle GW, Tatum EL (November 1941). “Genetic Control of Biochemical Reactions in Neurospora”. Proceedings of the National Academy of Sciences of the United States of America 27 (11): 499–506. Bibcode: 1941PNAS...27..499B. doi:10.1073/pnas.27.11.499. PMC 1078370. PMID 16588492.
- ^ Horowitz NH, Berg P, Singer M, Lederberg J, Susman M, Doebley J, Crow JF (January 2004). “A centennial: George W. Beadle, 1903-1989”. Genetics 166 (1): 1–10. doi:10.1534/genetics.166.1.1. PMC 1470705. PMID 15020400.
- ^ Judson HF (1996). The Eight Day of Creation (Expanded ed.). Plainview, NY (US): Cold Spring Harbor Laboratory Press
- ^ a b Watson JD (1965). Molecular Biology of the Gene. New York, NY, US: W.A. Benjamin, Inc.
- ^ Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD (1994). Molecular Biology of the Cell: Third Edition. London, UK: Garland Publishing, Inc.. ISBN 0-8153-1619-4
- ^ Moran LA, Horton HR, Scrimgeour KG, Perry MD (2012). Principles of Biochemistry: Fifth Edition. Upper Saddle River, NJ, US: Pearson
- ^ Lewin B (2004). Genes VIII. Upper Saddle River, NJ, US: Pearson/Prentice Hall
- ^ Piovesan A, Pelleri MC, Antonaros F, Strippoli P, Caracausi M, and Vitale L (2019). “On the length, weight and GC content of the human genome”. BMC Research Notes 12 (1): 106–173. doi:10.1186/s13104-019-4137-z. PMC 6391780. PMID 30813969.
- ^ Hubé F, and Francastel C (2015). “Mammalian Introns: When the Junk Generates Molecular Diversity”. International Journal of Molecular Sciences 16 (3): 4429–4452. doi:10.3390/ijms16034429. PMC 4394429. PMID 25710723.
- ^ Francis WR, and Wörheide G (2017). “Similar ratios of introns to intergenic sequence across animal genomes”. Genome Biology and Evolution 9 (6): 1582–1598. doi:10.1093/gbe/evx103. PMC 5534336. PMID 28633296.
- ^ Mortola E, Long M (2021). “Turning Junk into Us: How Genes Are Born”. American Scientist 109: 174–182.
- ^ Hopkin K (2009). “The Evolving Definition of a Gene: With the discovery that nearly all of the genome is transcribed, the definition of a "gene" needs another revision”. BioScience 59: 928–931. doi:10.1525/bio.2009.59.11.3.
- ^ Pearson H (2006). “What Is a Gene?”. Nature 441 (7092): 399–401. Bibcode: 2006Natur.441..398P. doi:10.1038/441398a. PMID 16724031.
- ^ Pennisi E (2007). “DNA study forces rethink of what it means to be a gene”. Science 316 (5831): 1556–1557. doi:10.1126/science.316.5831.1556. PMID 17569836.
- ^ Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV (November 2018). Eric Delwart, Luis Enjuanes. “Origins and Evolution of the Global RNA Virome”. mBio 9 (6): e02329–18. doi:10.1128/mBio.02329-18. PMC 6282212. PMID 30482837.
- ^ a b Pennisi E (June 2007). “Genomics. DNA study forces rethink of what it means to be a gene”. Science 316 (5831): 1556–7. doi:10.1126/science.316.5831.1556. PMID 17569836.
- ^ Marande W, Burger G (October 2007). “Mitochondrial DNA as a genomic jigsaw puzzle”. Science (AAAS) 318 (5849): 415. Bibcode: 2007Sci...318..415M. doi:10.1126/science.1148033. PMID 17947575.
- ^ Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigó R (January 2006). “Tandem chimerism as a means to increase protein complexity in the human genome”. Genome Research 16 (1): 37–44. doi:10.1101/gr.4145906. PMC 1356127. PMID 16344564.
- ^ a b c Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M (June 2007). “What is a gene, post-ENCODE? History and updated definition”. Genome Research 17 (6): 669–81. doi:10.1101/gr.6339607. PMID 17567988.
- ^ Noble D (September 2008). “Genes and causation”. Philosophical Transactions. Series A, Mathematical, Physical, and Engineering Sciences 366 (1878): 3001–15. Bibcode: 2008RSPTA.366.3001N. doi:10.1098/rsta.2008.0086. PMID 18559318.
- ^ “Blending Inheritance - an overview | ScienceDirect Topics”. 2021年7月3日閲覧。
- ^ "genesis". Oxford English Dictionary (3rd ed.). Oxford University Press. September 2005. (要購読、またはイギリス公立図書館への会員加入。)
- ^ Magner LN (2002). A History of the Life Sciences (Third ed.). Marcel Dekker, CRC Press. p. 371. ISBN 978-0-203-91100-6
- ^ Henig RM (2000). The Monk in the Garden: The Lost and Found Genius of Gregor Mendel, the Father of Genetics. Boston: Houghton Mifflin. pp. 1–9. ISBN 978-0395-97765-1
- ^ a b de Vries H (1889) (ドイツ語). Intracellulare Pangenese [Intracellular Pangenesis]. Jena: Verlag von Gustav Fischer Translated in 1908 from German to English by Open Court Publishing Co., Chicago, 1910
- ^ Bateson W (1906). “The progress of genetic research”. Report of the Third International Conference 1906 on Genetics. London, England: Royal Horticultural Society. pp. 90–97. "… the science itself [i.e. the study of the breeding and hybridisation of plants] is still nameless, and we can only describe our pursuit by cumbrous and often misleading periphrasis. To meet this difficulty I suggest for the consideration of this Congress the term Genetics, which sufficiently indicates that our labors are devoted to the elucidation of the phenomena of heredity and variation: in other words, to the physiology of Descent, with implied bearing on the theoretical problems of the evolutionist and the systematist, and application to the practical problems of breeders, whether of animals or plants."
- ^ Avery OT, Macleod CM, McCarty M (February 1944). “Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types : Induction of Transformation by a Desoxyribonucleic Acid Fraction Isolated From Pneumococcus Type III”. The Journal of Experimental Medicine 79 (2): 137–58. doi:10.1084/jem.79.2.137. PMC 2135445. PMID 19871359. Reprint: Avery OT, MacLeod CM, McCarty M (February 1979). “Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III”. The Journal of Experimental Medicine 149 (2): 297–326. doi:10.1084/jem.149.2.297. PMC 2184805. PMID 33226.
- ^ Hershey AD, Chase M (May 1952). “Independent functions of viral protein and nucleic acid in growth of bacteriophage”. The Journal of General Physiology 36 (1): 39–56. doi:10.1085/jgp.36.1.39. PMC 2147348. PMID 12981234.
- ^ Judson H (1979). The Eighth Day of Creation: Makers of the Revolution in Biology. Cold Spring Harbor Laboratory Press. pp. 51–169. ISBN 978-0-87969-477-7
- ^ Watson JD, Crick FH (April 1953). “Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid”. Nature 171 (4356): 737–8. Bibcode: 1953Natur.171..737W. doi:10.1038/171737a0. PMID 13054692.
- ^ Benzer S (June 1955). “Fine Structure of a Genetic Region in Bacteriophage”. Proceedings of the National Academy of Sciences of the United States of America 41 (6): 344–54. Bibcode: 1955PNAS...41..344B. doi:10.1073/pnas.41.6.344. PMC 528093. PMID 16589677.
- ^ Benzer S (November 1959). “On the Topology of the Genetic Fine Structure”. Proceedings of the National Academy of Sciences of the United States of America 45 (11): 1607–20. Bibcode: 1959PNAS...45.1607B. doi:10.1073/pnas.45.11.1607. PMC 222769. PMID 16590553.
- ^ Min Jou W, Haegeman G, Ysebaert M, Fiers W (May 1972). “Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein”. Nature 237 (5350): 82–8. Bibcode: 1972Natur.237...82J. doi:10.1038/237082a0. PMID 4555447.
- ^ Sanger F, Nicklen S, Coulson AR (December 1977). “DNA sequencing with chain-terminating inhibitors”. Proceedings of the National Academy of Sciences of the United States of America 74 (12): 5463–7. Bibcode: 1977PNAS...74.5463S. doi:10.1073/pnas.74.12.5463. PMC 431765. PMID 271968.
- ^ Adams JU (2008). “DNA Sequencing Technologies”. Nature Education Knowledge. SciTable (Nature Publishing Group) 1 (1): 193.
- ^ Huxley J (1942). Evolution: the Modern Synthesis. Cambridge, Massachusetts: MIT Press. ISBN 978-0262513661
- ^ Williams GC (2001). Adaptation and Natural Selection a Critique of Some Current Evolutionary Thought (Online ed.). Princeton: Princeton University Press. ISBN 9781400820108
- ^ Duret L (2008). “Neutral Theory: The Null Hypothesis of Molecular Evolution”. Nature Education 1: 218.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P『Molecular Biology of the Cell』(Fourth)Garland Science、New York、2002年、(#主要教科書の章番号を脚注リンク脇の数字 n.m に示す)。ISBN 978-0-8153-3218-3。
- ^ Stryer L, Berg JM, Tymoczko JL (2002). Biochemistry (5th ed.). San Francisco: W.H. Freeman. ISBN 978-0-7167-4955-4
- ^ Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, Cremer T (May 2005). “Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes”. PLOS Biology 3 (5): e157. doi:10.1371/journal.pbio.0030157. PMC 1084335. PMID 15839726.
- ^ Braig M, Schmitt CA (March 2006). “Oncogene-induced senescence: putting the brakes on tumor development”. Cancer Research 66 (6): 2881–4. doi:10.1158/0008-5472.CAN-05-4006. PMID 16540631.
- ^ a b Bennett PM (March 2008). “Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria”. British Journal of Pharmacology 153 (Suppl 1): S347-57. doi:10.1038/sj.bjp.0707607. PMC 2268074. PMID 18193080.
- ^ International Human Genome Sequencing Consortium (October 2004). “Finishing the euchromatic sequence of the human genome”. Nature 431 (7011): 931–45. Bibcode: 2004Natur.431..931H. doi:10.1038/nature03001. PMID 15496913.
- ^ a b Shafee, Thomas; Lowe, Rohan (2017). “Eukaryotic and prokaryotic gene structure”. WikiJournal of Medicine 4 (1). doi:10.15347/wjm/2017.002. ISSN 2002-4436.
- ^ Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (July 2008). “Mapping and quantifying mammalian transcriptomes by RNA-Seq”. Nature Methods 5 (7): 621–8. doi:10.1038/nmeth.1226. PMID 18516045.
- ^ Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G (April 2013). “Enhancers: five essential questions”. Nature Reviews. Genetics 14 (4): 288–95. doi:10.1038/nrg3458. PMC 4445073. PMID 23503198.
- ^ Maston GA, Evans SK, Green MR (2006). “Transcriptional regulatory elements in the human genome”. Annual Review of Genomics and Human Genetics 7: 29–59. doi:10.1146/annurev.genom.7.080505.115623. PMID 16719718.
- ^ Mignone F, Gissi C, Liuni S, Pesole G (2002-02-28). “Untranslated regions of mRNAs”. Genome Biology 3 (3): REVIEWS0004. doi:10.1186/gb-2002-3-3-reviews0004. PMC 139023. PMID 11897027.
- ^ Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ (December 2012). “Introns in UTRs: why we should stop ignoring them”. BioEssays 34 (12): 1025–34. doi:10.1002/bies.201200073. PMID 23108796.
- ^ Shkurin A, Pour SE, Hughes TR (April 2023). “Known sequence features explain half of all human gene ends”. NAR Genomics and Bioinformatics 5 (2): lqad031. doi:10.1093/nargab/lqad031. PMC 10072996. PMID 37035540.
- ^ Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J (June 2000). “Operons in Escherichia coli: genomic analyses and predictions”. Proceedings of the National Academy of Sciences of the United States of America 97 (12): 6652–7. Bibcode: 2000PNAS...97.6652S. doi:10.1073/pnas.110147297. PMC 18690. PMID 10823905.
- ^ Blumenthal T (November 2004). “Operons in eukaryotes”. Briefings in Functional Genomics & Proteomics 3 (3): 199–211. doi:10.1093/bfgp/3.3.199. PMID 15642184.
- ^ Jacob F, Monod J (June 1961). “Genetic regulatory mechanisms in the synthesis of proteins”. Journal of Molecular Biology 3 (3): 318–56. doi:10.1016/S0022-2836(61)80072-7. PMID 13718526.
- ^ Pozzoli U, Menozzi G, Comi GP, Cagliani R, Bresolin N, Sironi M (January 2007). “Intron size in mammals: complexity comes to terms with economy”. Trends in Genetics 23 (1): 20–24. doi:10.1016/j.tig.2006.10.003. PMID 17070957.
- ^ Marais G, Nouvellet P, Keightley PD, Charlesworth B (May 2005). “Intron size and exon evolution in Drosophila”. Genetics 170 (1): 481–485. doi:10.1534/genetics.104.037333. PMC 1449718. PMID 15781704.
- ^ Kumar A (September 2009). “An overview of nested genes in eukaryotic genomes”. Eukaryotic Cell 8 (9): 1321–1329. doi:10.1128/EC.00143-09. PMC 2747821. PMID 19542305..
- ^ Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA (June 2005). “Interchromosomal associations between alternatively expressed loci”. Nature 435 (7042): 637–645. Bibcode: 2005Natur.435..637S. doi:10.1038/nature03574. PMID 15880101.
- ^ Williams A, Spilianakis CG, Flavell RA (April 2010). “Interchromosomal association and gene regulation in trans”. Trends in Genetics 26 (4): 188–197. doi:10.1016/j.tig.2010.01.007. PMC 2865229. PMID 20236724.
- ^ Lei Q, Li C, Zuo Z, Huang C, Cheng H, Zhou R (March 2016). “Evolutionary Insights into RNA trans-Splicing in Vertebrates”. Genome Biology and Evolution 8 (3): 562–577. doi:10.1093/gbe/evw025. PMC 4824033. PMID 26966239.
- ^ Wright BW, Molloy MP, Jaschke PR (March 2022). “Overlapping genes in natural and engineered genomes”. Nature Reviews. Genetics 23 (3): 154–168. doi:10.1038/s41576-021-00417-w. PMC 8490965. PMID 34611352.
- ^ a b Eddy SR (December 2001). “Non-coding RNA genes and the modern RNA world”. Nature Reviews. Genetics 2 (12): 919–29. doi:10.1038/35103511. PMID 11733745.
- ^ Crick FH, Barnett L, Brenner S, Watts-Tobin RJ (December 1961). “General nature of the genetic code for proteins”. Nature 192 (4809): 1227–32. Bibcode: 1961Natur.192.1227C. doi:10.1038/1921227a0. PMID 13882203.
- ^ Crick FH (October 1962). “The genetic code”. Scientific American (WH Freeman and Company) 207 (4): 66–74. Bibcode: 1962SciAm.207d..66C. doi:10.1038/scientificamerican1062-66. PMID 13882204.
- ^ Woodson SA (May 1998). “Ironing out the kinks: splicing and translation in bacteria”. Genes & Development 12 (9): 1243–7. doi:10.1101/gad.12.9.1243. PMID 9573040.
- ^ Jacob F, Monod J (June 1961). “Genetic regulatory mechanisms in the synthesis of proteins”. Journal of Molecular Biology 3 (3): 318–56. doi:10.1016/S0022-2836(61)80072-7. PMID 13718526.
- ^ Koonin EV, Dolja VV (January 1993). “Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences”. Critical Reviews in Biochemistry and Molecular Biology 28 (5): 375–430. doi:10.3109/10409239309078440. PMID 8269709.
- ^ Domingo E (2001). “RNA Virus Genomes”. eLS. doi:10.1002/9780470015902.a0001488.pub2. ISBN 978-0470016176.
- ^ Domingo E, Escarmís C, Sevilla N, Moya A, Elena SF, Quer J, Novella IS, Holland JJ (June 1996). “Basic concepts in RNA virus evolution”. FASEB Journal 10 (8): 859–64. doi:10.1096/fasebj.10.8.8666162. PMID 8666162.
- ^ Miko I (2008). “Gregor Mendel and the Principles of Inheritance”. Nature Education Knowledge. SciTable (Nature Publishing Group) 1 (1): 134.
- ^ Chial H (2008). “Mendelian Genetics: Patterns of Inheritance and Single-Gene Disorders”. Nature Education Knowledge. SciTable (Nature Publishing Group) 1 (1): 63.
- ^ McCarthy D, Minner C, Bernstein H, Bernstein C (October 1976). “DNA elongation rates and growing point distributions of wild-type phage T4 and a DNA-delay amber mutant”. Journal of Molecular Biology 106 (4): 963–81. doi:10.1016/0022-2836(76)90346-6. PMID 789903.
- ^ a b Lobo I, Shaw K (2008). “Discovery and Types of Genetic Linkage”. Nature Education Knowledge. SciTable (Nature Publishing Group) 1 (1): 139.
- ^ Nachman MW, Crowell SL (September 2000). “Estimate of the mutation rate per nucleotide in humans”. Genetics 156 (1): 297–304. doi:10.1093/genetics/156.1.297. PMC 1461236. PMID 10978293.
- ^ Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ (April 2010). “Analysis of genetic inheritance in a family quartet by whole-genome sequencing”. Science 328 (5978): 636–9. Bibcode: 2010Sci...328..636R. doi:10.1126/science.1186802. PMC 3037280. PMID 20220176.
- ^ Drake JW, Charlesworth B, Charlesworth D, Crow JF (April 1998). “Rates of spontaneous mutation”. Genetics 148 (4): 1667–86. doi:10.1093/genetics/148.4.1667. PMC 1460098. PMID 9560386.
- ^ Pyeritz, Reed E., Bruce R. Korf, and Wayne W. Grody, eds. Emery and Rimoin’s principles and practice of medical genetics and genomics: foundations. Academic Press, 2018.
- ^ “What kinds of gene mutations are possible?”. Genetics Home Reference. United States National Library of Medicine (2015年5月11日). 2015年5月19日閲覧。
- ^ Andrews CA (2010). “Natural Selection, Genetic Drift, and Gene Flow Do Not Act in Isolation in Natural Populations”. Nature Education Knowledge. SciTable (Nature Publishing Group) 3 (10): 5.
- ^ Patterson C (November 1988). “Homology in classical and molecular biology”. Molecular Biology and Evolution 5 (6): 603–25. doi:10.1093/oxfordjournals.molbev.a040523. PMID 3065587.
- ^ Graur D (2016). Molecular and Genome Evolution. Sunderland MA (US): Sinauer Associates, Inc.. ISBN 9781605354699
- ^ Graur D (2016). Molecular and Genome Evolution. Sunderland MA (US): Sinauer Associates, Inc.. ISBN 9781605354699
- ^ Jensen RA (2001). “Orthologs and paralogs - we need to get it right”. Genome Biology 2 (8): INTERACTIONS1002. doi:10.1186/gb-2001-2-8-interactions1002. PMC 138949. PMID 11532207.
- ^ Studer RA, Robinson-Rechavi M (May 2009). “How confident can we be that orthologs are similar, but paralogs differ?”. Trends in Genetics 25 (5): 210–6. doi:10.1016/j.tig.2009.03.004. PMID 19368988.
- ^ Altenhoff AM, Studer RA, Robinson-Rechavi M, Dessimoz C (2012). “Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs”. PLOS Computational Biology 8 (5): e1002514. Bibcode: 2012PLSCB...8E2514A. doi:10.1371/journal.pcbi.1002514. PMC 3355068. PMID 22615551.
- ^ a b Guerzoni D, McLysaght A (November 2011). “De novo origins of human genes”. PLOS Genetics 7 (11): e1002381. doi:10.1371/journal.pgen.1002381. PMC 3213182. PMID 22102832.
- ^ Reams AB, Roth JR (February 2015). “Mechanisms of gene duplication and amplification”. Cold Spring Harbor Perspectives in Biology 7 (2): a016592. doi:10.1101/cshperspect.a016592. PMC 4315931. PMID 25646380.
- ^ Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW (December 2006). “The evolution of mammalian gene families”. PLOS ONE 1 (1): e85. Bibcode: 2006PLoSO...1...85D. doi:10.1371/journal.pone.0000085. PMC 1762380. PMID 17183716.
- ^ Knowles DG, McLysaght A (October 2009). “Recent de novo origin of human protein-coding genes”. Genome Research 19 (10): 1752–9. doi:10.1101/gr.095026.109. PMC 2765279. PMID 19726446.
- ^ Wu DD, Irwin DM, Zhang YP (November 2011). “De novo origin of human protein-coding genes”. PLOS Genetics 7 (11): e1002379. doi:10.1371/journal.pgen.1002379. PMC 3213175. PMID 22102831.
- ^ McLysaght A, Guerzoni D (September 2015). “New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation”. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 370 (1678): 20140332. doi:10.1098/rstb.2014.0332. PMC 4571571. PMID 26323763.
- ^ Neme R, Tautz D (February 2013). “Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution”. BMC Genomics 14 (1): 117. doi:10.1186/1471-2164-14-117. PMC 3616865. PMID 23433480.
- ^ Treangen TJ, Rocha EP (January 2011). “Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes”. PLOS Genetics 7 (1): e1001284. doi:10.1371/journal.pgen.1001284. PMC 3029252. PMID 21298028.
- ^ Ochman H, Lawrence JG, Groisman EA (May 2000). “Lateral gene transfer and the nature of bacterial innovation”. Nature 405 (6784): 299–304. Bibcode: 2000Natur.405..299O. doi:10.1038/35012500. PMID 10830951.
- ^ Keeling PJ, Palmer JD (August 2008). “Horizontal gene transfer in eukaryotic evolution”. Nature Reviews. Genetics 9 (8): 605–18. doi:10.1038/nrg2386. PMID 18591983.
- ^ Schönknecht G, Chen WH, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Bräutigam A, Baker BJ, Banfield JF, Garavito RM, Carr K, Wilkerson C, Rensing SA, Gagneul D, Dickenson NE, Oesterhelt C, Lercher MJ, Weber AP (March 2013). “Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote”. Science 339 (6124): 1207–10. Bibcode: 2013Sci...339.1207S. doi:10.1126/science.1231707. PMID 23471408.
- ^ Ridley, M. (2006). Genome. New York, NY: Harper Perennial. ISBN 0-06-019497-9
- ^ Banerjee S, Bhandary P, Woodhouse M, Sen TZ, Wise RP, Andorf CM (Apr 2021). “FINDER: an automated software package to annotate eukaryotic genes from RNA-Seq data and associated protein sequences”. BMC Bioinformatics 44 (9): e89. doi:10.1186/s12859-021-04120-9. PMC 8056616. PMID 33879057.
- ^ Watson, JD, Baker TA, Bell SP, Gann A, Levine M, Losick R. (2004). "Ch9-10", Molecular Biology of the Gene, 5th ed., Peason Benjamin Cummings; CSHL Press.
- ^ “Integr8 – A.thaliana Genome Statistics”. 2015年4月26日閲覧。
- ^ “Understanding the Basics”. The Human Genome Project. 2015年4月26日閲覧。
- ^ “WS227 Release Letter”. WormBase (2011年8月10日). 2013年11月28日時点のオリジナルよりアーカイブ。2013年11月19日閲覧。
- ^ a b Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Huang X, Li W, Li J, Liu Z, Li L, Liu J, Qi Q, Liu J, Li L, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Zhang J, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Ren X, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Wang J, Zhao W, Li P, Chen W, Wang X, Zhang Y, Hu J, Wang J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Li G, Liu S, Tao M, Wang J, Zhu L, Yuan L, Yang H (April 2002). “A draft sequence of the rice genome (Oryza sativa L. ssp. indica)”. Science 296 (5565): 79–92. Bibcode: 2002Sci...296...79Y. doi:10.1126/science.1068037. PMID 11935017.
- ^ Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (April 1981). “Sequence and organization of the human mitochondrial genome”. Nature 290 (5806): 457–65. Bibcode: 1981Natur.290..457A. doi:10.1038/290457a0. PMID 7219534.
- ^ Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC (March 2000). “The genome sequence of Drosophila melanogaster”. Science 287 (5461): 2185–95. Bibcode: 2000Sci...287.2185.. doi:10.1126/science.287.5461.2185. PMID 10731132.
- ^ Pertea M, Salzberg SL (2010). “Between a chicken and a grape: estimating the number of human genes”. Genome Biology 11 (5): 206. doi:10.1186/gb-2010-11-5-206. PMC 2898077. PMID 20441615.
- ^ Belyi VA, Levine AJ, Skalka AM (December 2010). “Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the parvoviridae and circoviridae are more than 40 to 50 million years old”. Journal of Virology 84 (23): 12458–62. doi:10.1128/JVI.01789-10. PMC 2976387. PMID 20861255.
- ^ Flores R, Di Serio F, Hernández C (February 1997). “Viroids: The Noncoding Genomes”. Seminars in Virology 8 (1): 65–73. doi:10.1006/smvy.1997.0107.
- ^ Zonneveld BJ (2010). “New Record Holders for Maximum Genome Size in Eudicots and Monocots”. Journal of Botany 2010: 1–4. doi:10.1155/2010/527357.
- ^ Perez-Iratxeta C, Palidwor G, Andrade-Navarro MA (December 2007). “Towards completion of the Earth's proteome”. EMBO Reports 8 (12): 1135–41. doi:10.1038/sj.embor.7401117. PMC 2267224. PMID 18059312.
- ^ Muller HJ (1966). “The gene material as the initiator and the organizing basis of life”. American Naturalist 100 (915): 493–517. doi:10.1086/282445. JSTOR 2459205.
- ^ Ohno S (1972). “So much "junk" DNA in our genome”. Brookhaven Symposia in Biology 23: 366–370. PMID 5065367.
- ^ Hatje K, Mühlhausen S, Simm D, Killmar M (2019). “The Protein-Coding Human Genome: Annotating High-Hanging Fruits.”. BioEssays 41 (11): 1900066. doi:10.1002/bies.201900066. PMID 31544971.
- ^ Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, White RE, Rodriguez-Tomé P, Aggarwal A, Bajorek E, Bentolila S, Birren BB, Butler A, Castle AB, Chiannilkulchai N, Chu A, Clee C, Cowles S, Day PJ, Dibling T, Drouot N, Dunham I, Duprat S, East C, Edwards C, Fan JB, Fang N, Fizames C, Garrett C, Green L, Hadley D, Harris M, Harrison P, Brady S, Hicks A, Holloway E, Hui L, Hussain S, Louis-Dit-Sully C, Ma J, MacGilvery A, Mader C, Maratukulam A, Matise TC, McKusick KB, Morissette J, Mungall A, Muselet D, Nusbaum HC, Page DC, Peck A, Perkins S, Piercy M, Qin F, Quackenbush J, Ranby S, Reif T, Rozen S, Sanders C, She X, Silva J, Slonim DK, Soderlund C, Sun WL, Tabar P, Thangarajah T, Vega-Czarny N, Vollrath D, Voyticky S, Wilmer T, Wu X, Adams MD, Auffray C, Walter NA, Brandon R, Dehejia A, Goodfellow PN, Houlgatte R, Hudson JR, Ide SE, Iorio KR, Lee WY, Seki N, Nagase T, Ishikawa K, Nomura N, Phillips C, Polymeropoulos MH, Sandusky M, Schmitt K, Berry R, Swanson K, Torres R, Venter JC, Sikela JM, Beckmann JS, Weissenbach J, Myers RM, Cox DR, James MR, Bentley D, Deloukas P, Lander ES, Hudson TJ (October 1996). “A gene map of the human genome”. Science 274 (5287): 540–6. Bibcode: 1996Sci...274..540S. doi:10.1126/science.274.5287.540. PMID 8849440.
- ^ Chi KR (October 2016). “The dark side of the human genome” (英語). Nature 538 (7624): 275–277. Bibcode: 2016Natur.538..275C. doi:10.1038/538275a. PMID 27734873.
- ^ Ezkurdia, Iakes; Juan, David; Rodriguez, Jose Manuel; Frankish, Adam; Diekhans, Mark; Harrow, Jennifer; Vazquez, Jesus; Valencia, Alfonso et al. (2014-11-15). “Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes” (英語). Human Molecular Genetics 23 (22): 5866–5878. doi:10.1093/hmg/ddu309. ISSN 0964-6906. PMC PMC4204768. PMID 24939910.
- ^ “Human assembly and gene annotation”. Ensembl (2022年). 2023年2月28日閲覧。
- ^ a b Hutchison CA, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi ZQ, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC (March 2016). “Design and synthesis of a minimal bacterial genome”. Science 351 (6280): aad6253. Bibcode: 2016Sci...351.....H. doi:10.1126/science.aad6253. PMID 27013737.
- ^ Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, Smith HO, Venter JC (January 2006). “Essential genes of a minimal bacterium”. Proceedings of the National Academy of Sciences of the United States of America 103 (2): 425–30. Bibcode: 2006PNAS..103..425G. doi:10.1073/pnas.0510013103. PMC 1324956. PMID 16407165.
- ^ Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D'Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabási AL, Oltvai ZN, Osterman AL (October 2003). “Experimental determination and system level analysis of essential genes in Escherichia coli MG1655”. Journal of Bacteriology 185 (19): 5673–84. doi:10.1128/jb.185.19.5673-5684.2003. PMC 193955. PMID 13129938.
- ^ Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006). “Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection”. Molecular Systems Biology 2: 2006.0008. doi:10.1038/msb4100050. PMC 1681482. PMID 16738554.
- ^ a b Juhas M, Reuß DR, Zhu B, Commichau FM (November 2014). “Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering”. Microbiology 160 (Pt 11): 2341–2351. doi:10.1099/mic.0.079376-0. PMID 25092907.
- ^ Tu Z, Wang L, Xu M, Zhou X, Chen T, Sun F (February 2006). “Further understanding human disease genes by comparing with housekeeping genes and other genes”. BMC Genomics 7: 31. doi:10.1186/1471-2164-7-31. PMC 1397819. PMID 16504025.
- ^ Georgi B, Voight BF, Bućan M (May 2013). “From mouse to human: evolutionary genomics analysis of human orthologs of essential genes”. PLOS Genetics 9 (5): e1003484. doi:10.1371/journal.pgen.1003484. PMC 3649967. PMID 23675308.
- ^ Eisenberg E, Levanon EY (October 2013). “Human housekeeping genes, revisited”. Trends in Genetics 29 (10): 569–74. doi:10.1016/j.tig.2013.05.010. PMID 23810203.
- ^ Amsterdam A, Hopkins N (September 2006). “Mutagenesis strategies in zebrafish for identifying genes involved in development and disease”. Trends in Genetics 22 (9): 473–8. doi:10.1016/j.tig.2006.06.011. PMID 16844256.
- ^ “About the HGNC”. HGNC Database of Human Gene Names. HUGO Gene Nomenclature Committee. 2015年5月14日閲覧。
- ^ Cohen SN, Chang AC (May 1973). “Recircularization and autonomous replication of a sheared R-factor DNA segment in Escherichia coli transformants”. Proceedings of the National Academy of Sciences of the United States of America 70 (5): 1293–7. Bibcode: 1973PNAS...70.1293C. doi:10.1073/pnas.70.5.1293. PMC 433482. PMID 4576014.
- ^ Esvelt KM, Wang HH (2013). “Genome-scale engineering for systems and synthetic biology”. Molecular Systems Biology 9 (1): 641. doi:10.1038/msb.2012.66. PMC 3564264. PMID 23340847.
- ^ Tan WS, Carlson DF, Walton MW, Fahrenkrug SC, Hackett PB (2012). “Precision editing of large animal genomes”. Advances in Genetics Volume 80. 80. pp. 37–97. doi:10.1016/B978-0-12-404742-6.00002-8. ISBN 9780124047426. PMC 3683964. PMID 23084873
- ^ Puchta H, Fauser F (2013). “Gene targeting in plants: 25 years later”. The International Journal of Developmental Biology 57 (6–8): 629–37. doi:10.1387/ijdb.130194hp. PMID 24166445.
- ^ Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (November 2013). “Genome engineering using the CRISPR-Cas9 system”. Nature Protocols 8 (11): 2281–2308. doi:10.1038/nprot.2013.143. PMC 3969860. PMID 24157548.
- ^ Kittleson JT, Wu GC, Anderson JC (August 2012). “Successes and failures in modular genetic engineering”. Current Opinion in Chemical Biology 16 (3–4): 329–36. doi:10.1016/j.cbpa.2012.06.009. PMID 22818777.
- ^ Berg P, Mertz JE (January 2010). “Personal reflections on the origins and emergence of recombinant DNA technology”. Genetics 184 (1): 9–17. doi:10.1534/genetics.109.112144. PMC 2815933. PMID 20061565.
- ^ Guan C, Ye C, Yang X, Gao J (February 2010). “A review of current large-scale mouse knockout efforts”. Genesis 48 (2): 73–85. doi:10.1002/dvg.20594. PMID 20095055.
- ^ Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melchner H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B (September 2004). “The knockout mouse project”. Nature Genetics 36 (9): 921–4. doi:10.1038/ng0904-921. PMC 2716027. PMID 15340423.
- ^ Deng C (October 2007). “In celebration of Dr. Mario R. Capecchi's Nobel Prize”. International Journal of Biological Sciences 3 (7): 417–9. doi:10.7150/ijbs.3.417. PMC 2043165. PMID 17998949.
Gene
出典: フリー百科事典『ウィキペディア(Wikipedia)』 (2021/06/26 05:15 UTC 版)
「アメリカ国立生物工学情報センター」の記事における「Gene」の解説
Geneデータベースは、遺伝子に関する情報を特徴付けて整理するために、NCBIで実装されている。配列情報、タンパク質機能、構造、ゲノム上の位置、発現、および配列類似性に関する情報を蒐集している。一意のGeneIDが各遺伝子レコードに割り当てられており、改訂の経歴をたどることができる。既知または予測された遺伝子のレコードがここで確立され、マップ位置やヌクレオチド配列によって画定されている。Geneには、NCBIの他のデータベースとの統合の改善、分類範囲の拡大、Entrezシステムによって提供されるクエリと検索の拡張オプションなど、前身のLocusLinkに比べていくつかの利点がある。
※この「Gene」の解説は、「アメリカ国立生物工学情報センター」の解説の一部です。
「Gene」を含む「アメリカ国立生物工学情報センター」の記事については、「アメリカ国立生物工学情報センター」の概要を参照ください。
- Geneのページへのリンク